X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
K. Finkelstein (July 12th, 2006)
CHESS, Cornell University, Ithaca, NY 14853
Although scientists know much about the static structures of DNA, RNA and proteins, new tools are needed to explore the dynamics of these complex molecules. Hoping to provide missing data to test current theories and models, the Pollack group in the School of Applied and Engineering Physics at Cornell University has recently shown that careful x-ray measurements can be used to get quantitative information about the electrostatic interactions between DNA molecules in solution. Their work was published recently in a Physical Review Letters article entitled “Measuring Inter-DNA Potentials in Solution".
Nucleic acids are a very important class of biological molecules. DNAs encode the genetic blueprint of living matter; RNAs have roles such as messengers and transporters, and the existence of enzymatic RNAs supports the hypothesis of a primordial RNA world. Nucleic acids also exhibit a wide range of complex dynamics. For example: the compaction of long DNA strands in the nucleus and the folding of some RNAs into catalytically active 3D structures. Quantitative understanding of nucleic acids structure and dynamics requires quantitative knowledge of the forces and underlying physical mechanisms that govern the energetics and interactions of these materials.
Electrostatic interactions are essential to nucleic acids due to their remarkably high charge density (-1e/1.7Å for dsDNA). Strong like-charge repulsion is expected between nucleic acids, but charged salt ions in solution can screen repulsion and influence the electrostatic collapse in the early events during RNA folding. It is well-known that the valence of positively charged counterions has dramatic effects on the interaction between nucleic acid molecules. With rather dilute trivalent or higher valence counterions, DNAs attract each other and can condense into aggregates. Physical understanding of nucleic acids electrostatics is first based on mean field Poisson-Boltzmann equations that fail to account for like-charge attraction. Extensive theoretical work that considers the discrete charge and spatial correlation of counterions has helped in understanding this puzzle, but few quantitative measurements of the nucleic acids interactions are available to allow direct comparisons between theory and experiment.
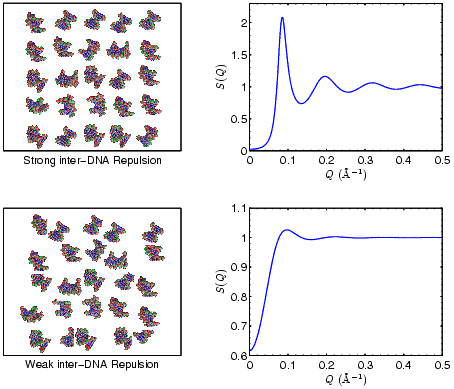
Figure 1: Demonstration of
the relationship between the spatial correlations of
DNA molecules in solution and the x-ray scattering
structure factor S(Q).
Postdoctoral fellow Xiangyun Qiu and graduate students Lisa W. Kwok, Hye Yoon Park, Jessica S. Lamb, and Kurt Andresen have shown that x-rays can be used to measure the forces between short strands of dsDNAs (25 base pairs) in solution. Under low salt concentration DNAs try to avoid each other and form an ordered lattice. With increasing concentration and at higher ion valence the inter-DNA force turns attractive. The spatial arrangements of DNAs were measured by small-angle x-ray scattering (SAXS) carried out at the C1 station at the Cornell High Energy Synchrotron Source (CHESS). The inter-DNA forces are quantified via straightforward analysis of the total SAXS profiles in conjunction with accepted theoretical models of polyelectrolytes. By varying the counterion valence and concentration, they “tuned” the inter-DNA interactions from repulsive to weakly attractive. Repulsive interactions dominate up to moderate monovalent ion concentrations, while weak attractions are observed at surprisingly low divalent ion concentrations. The quantitative results are found to differ significantly from predictions of available analytical theories.
Measuring inter-DNA potentials in solution provides an important experimental guide to understanding nucleic acids interactions. Determined interaction parameters can direct numeric simulations describing the repulsion and attraction of like charged polyelectrolytes. This SAXS x-ray method shows that readily available techniques can answer questions about dynamics in biological systems.
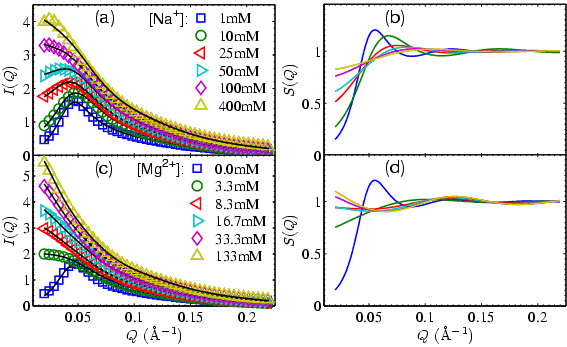
Figure 2: Experiments SAXS
profiles showing the tuning of inter-DNA forces with DNA concentrations
around 0.6mM. (a) and (c) show raw scattering curves
offset for clarity. I(Q)=S(Q)*P(Q) where P(Q) is the
molecular form factor. The dark lines are fits. (b)
and (d) show S(Q)s corresponding to (a) and (c).
Weakening of the inter-DNA repulsion is indicated with
increasing salt concentrations. The upturn in S(Q) for
[Mg2+] > 16mM is evidence for attraction.
Recent Publication:
X. Qui, L. W. Kwok, H.-Y. Park, J. S. Lamb, K. Andresen, and L. Pollack; "Measuring Inter-DNA Potentials in Solution", Physical Review Letters, 96, 138101 (2006).
