X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
A receptor-modifying demainase in complex with a signaling phosphatase reveals reciprocal regulation.
X. Chao, T.J. Muff, S.-Y. Park, S. Zhang, A.M. Pollard, G.W. Ordal, A.M. Bilwes, and B.R. Crane
Cell (2006), Vol. 124, 561-571
Bacteria swim towards attractants such as nutrients and away from repellants such as toxins. This process, chemotaxis, requires sensing the concentration of a chemical in the environment and transmitting a signal to the bacterial flagellum to modify the bacterium's swimming behavior. Major proteins involved are MCP (a transmembrane signal receptor), CheA (receives the signal from MCP and phosphorylates CheY), CheY (binds to the flagellar switch complex to control the direction in which the flagellum rotates), CheC and CheD (terminate the signal by modifying MCP), CheA and CheR (regulate sensitivity of MCP to avoid signal saturation).
This study focuses on the interaction between CheC and CheD. A crystal structure of a CheC:CheD complex was determined to 2.4 Å, using data collected at the the CHESS F2 station and phasing by molecular replacement with a CheC model. Additionally, the activity of various CheD mutants was measured to pinpoint the importance of specific amino acids to enzyme function.
The CheD structure is similar to that of the CNF1 toxin, which functions by deamidating a critical Gln residue in a signaling protein; this suggests the mechanism by which CheD modifies MCP. CheC contains a recognition motif similar to that in the MCP receptor, and the CheC:CheD complex mimics the receptor:CheD interaction (see the figure). When the complex is formed, CheC inhibits CheD, while CheD stimulates the phosphatase activity of CheC. Mutation of the central residue in the CheC binding region to Gln causes CheC to become a substrate instead of an inhibitor of CheD, confirming the importance of the Gln in the proposed CheD-binding region of MCP. Another mutation drastically reduces CheC-CheD association and impairs chemotaxis in vivo, consistent with the importance of the CheC:CheD complex. Phosphorylated CheY, whose phosphate group is removed by CheC to terminate signaling, stabilizes the CheC:CheD complex and correspondingly reduces availability of CheD
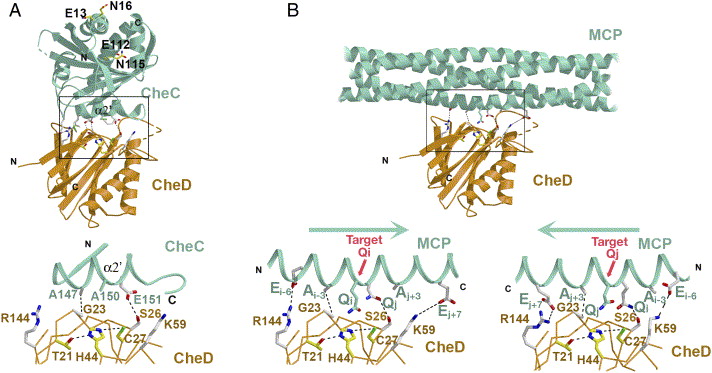
(A) CheC (green) inserts its α2′ helix into the CheD
active center (orange) and blocks the catalytic center
(yellow side chains). The expanded view of the boxed
interface region shows interactions (dotted lines)
between key side chains (S26-to-E151, A147-to-G23,
white) that mediate the contact. CheC A150 resides at
the position of the substrate Gln in receptors.
(B) Proposed interaction between CheD and receptor MCP1143C,
based on the structures of the CheD:CheC complex and
the receptor. CheC A150 is replaced by a substrate Gln.
The symmetry of the receptor substrate likely allows
CheD to bind to the helix in either of two
orientations, so that S26 can interact with either Qi or Qj.
Also see:
Complete article here (pdf)
online: 02/23/2006
