X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
Structure of a DNA glycosylase searching for lesions.
A. Banerjee, W.L. Santos, and G.L. Verdine
Science (2006), Vol. 311, 1153-1157
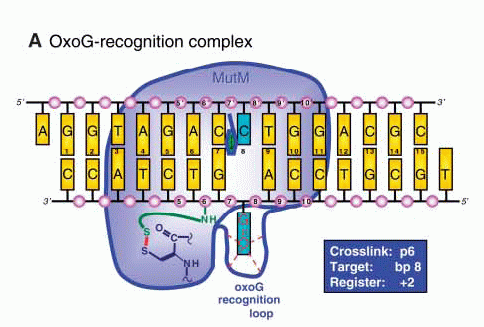
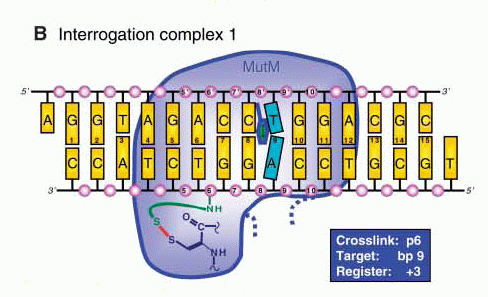
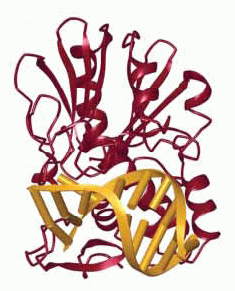
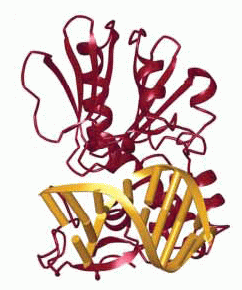
Structures of (A) the lesion recognition complex (LRC)
formed between MutM and DNA containing an
8-oxo-guanine nucleotide and (B) an interrogation
complex (IC) formed between MutM and undamaged DNA;
complexes are stabilized by disulfide cross-linking
between protein and DNA. Structures are shown
schematically above and in a ribbon form below. Note
differences in the lower left of the figure, where the
oxoG base is extruded from the DNA helix into a
binding pocket in MutM, while the corresponding normal
base remains intrahelical.
DNA glycosylases, such as MutM in bacteria and OGG1 in mammals, have the difficult task of scanning millions of base pairs of DNA to locate a few damaged nucleotides, which may differ only slightly from normal ones, and removing them. The scanning process is highly efficient, requiring no use of biochemical energy, and how it works has been a mystery.
A crystal structure of a "lesion recognition complex (LRC)" of MutM with DNA containing a modified guanine residue (oxoG) revealed that a nucleotide identified as damaged is extruded from the DNA double helix into an active-site pocket on the enzyme, where it can be clipped out (Fromme & Verdine, J. Biol. Chem. (2003) 278, 51543). This does not, however, tell us how MutM checks a long stretch of DNA for damage; extrusion of each residue for interrogation seems unlikely to be sufficiently efficient.
Verdine and colleagues have now used a disulfide cross-linking technique to trap MutM in the process of interrogating undamaged DNA. Structures were determined for 3 different complexes, varying in DNA sequence and attachment point of the disulfide link. All showed insertion of the sidechain of MutM residue Phe 114 into the DNA helix; the adjacent base pair is buckled but intact. In the LRC, the Phe 114 sidechain is joined by those of Arg 112 and Met 77, with concomitant bending of the DNA and extrusion of the oxoG residue. The authors suggest that the buckling of a base pair, caused by the insertion of the aromatic ring of Phe 114, destabilizes it, so that an abnormal pairing will be broken and the damaged base extruded for removal. In the case of MutM, the active site is also quite specific for oxoG, so that a normal base extruded by mistake will not be removed.
Also see:
Complete article here (pdf)
online: 04/10/2006
