X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
Contact: M. Szebenyi (dms35@cornell.edu)
Structurally mapping the diverse phenotype
of adeno-associated virus serotype 4.
L. Govindasamy, W. Padron, R. McKenna, N. Muzyczka, N. Kaludov,
J. A. Chiorini, and M. Agbandje-McKenna
J. Virology (2006), Vol. 80, 11556-11570
Adeno-associated virus (AAV) is potentially useful as a carrier of DNA to correct genetic defects; numerous animal experiments using an AAV vector have been carried out, and human trials are beginning. For best results with minimal side effects, corrective genes should be delivered only to the appropriate cells. This targeting is controlled by engineering the viral surface of AAV - but what surface changes are needed? Luckily, a number of AAV serotypes exist, with different cell binding and antigenic characteristics, and determination of their structures allows correlation of surface detail with infectivities.
The Agbandje-McKenna group, at U. Florida, has determined a number of AAV structures. The most recent is AAV4, whose structure was determined by molecular replacement, using data from CHESS F1 station. Comparison of the new structure with that of AAV2 (determined earlier by another CHESS user, the Chapman group) revealed that the core regions of the viral capsid proteins (VPs), and the packing of VPs on the viral surface, are essentially the same in the different serotypes, while the shapes, and charges, of surface features differ substantially (see the figure). Specific amino acids involved in receptor recognition have been identified, as well as locations in which loops could be inserted or deleted without affecting VP packing; this information will be very helpful in guiding genetic engineering of AAVs.
The AAV4 structure also contained bound dAMP, a DNA nucleotide, in a pocket on the inside of the capsid. This was unexpected - amino acids making up the dAMP binding pocket are conserved among different AAVs (indicating some importance to the structure), but DNA binding to related viruses such as canine parvovirus does not involve the equivalent region. Further experiments will be needed to solve the mystery.
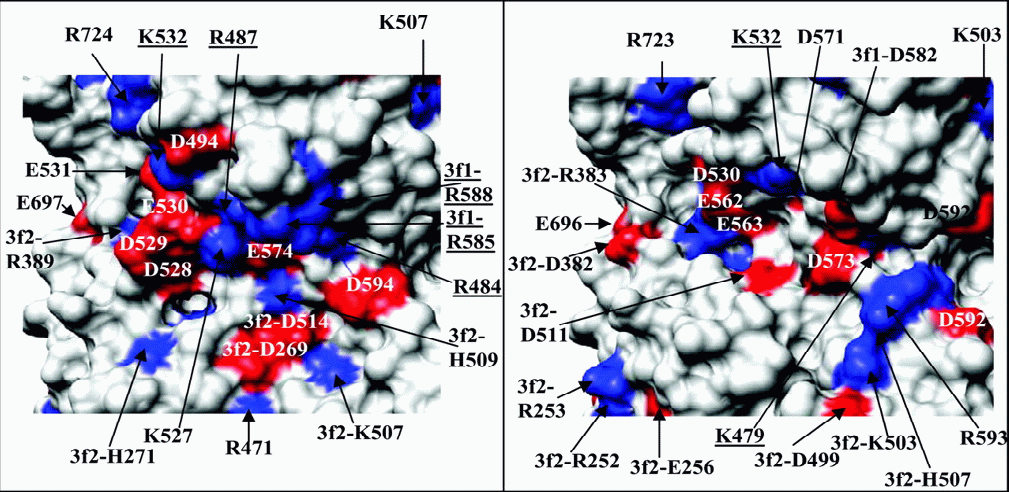
Details
of the surface structure in the heparin binding region
of AAV2 (left) and the corresponding region of AAV4
(right). Charged residues (basic in blue, acidic in
red) are labeled; those involved in binding heparin
are underlined.
