X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
This past March MacCHESS offered an introductory minicourse, BioSAXS Essentials, designed to cover the basics of small-angle-x-ray solution scattering from biological solutions. Demand for the minicourse immediately exceeded expectations, so the course was repeated in May with a class size twice that of the March offering.
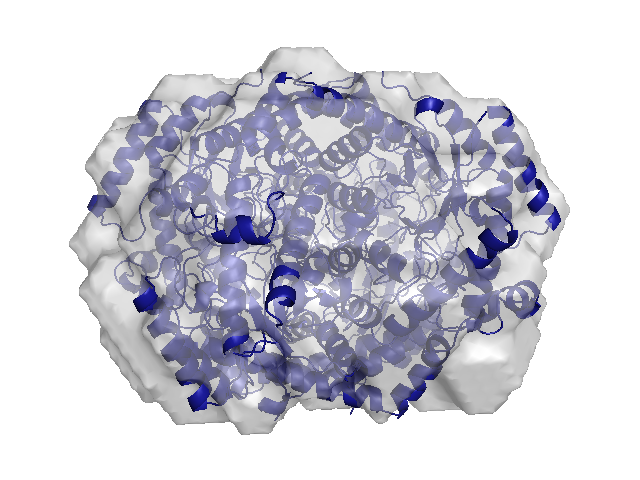
BioSAXS is rapidly becoming popular with molecular biologists due to its ability to produce structural information from a wide range of solution conditions without the need for growing crystals. In the classic BioSAXS experiment, a small volume of protein solution (~10 microliters) is exposed to a well-collimated x-ray beam with energy in the range of 8-12 keV. Scattering intensity at small angles (less than 5 degrees) is collected on an x-ray area detector under very low-background noise conditions and integrated to yield a one-dimensional intensity profile. Compared to crystallography, BioSAXS is a low-resolution technique, but advances in algorithms have demonstrated that scattering profiles contain far more detailed information than previously thought.
Structural biology has shifted focus to large multi-component complexes, molecules with intrinsic disorder, domains with flexible linkers, and other challenging systems that are often beyond the reach of current crystallographic methods. BioSAXS plays a unique role in biology quite different than that of crystallography. The method is used more in conjunction with other biophysical techniques and it applies much earlier in the research process. In addition to answering questions about the basic size, shape, and oligomeric state in solution, the method also yields information about flexibility, unfolding, and interparticle interactions. It can help distinguish between competing structural models. It has been combined with NMR data and atomistic simulations, and used to determine spatial distributions of domains with flexible linkers. Novel uses for the data are appearing regularly in the literature.
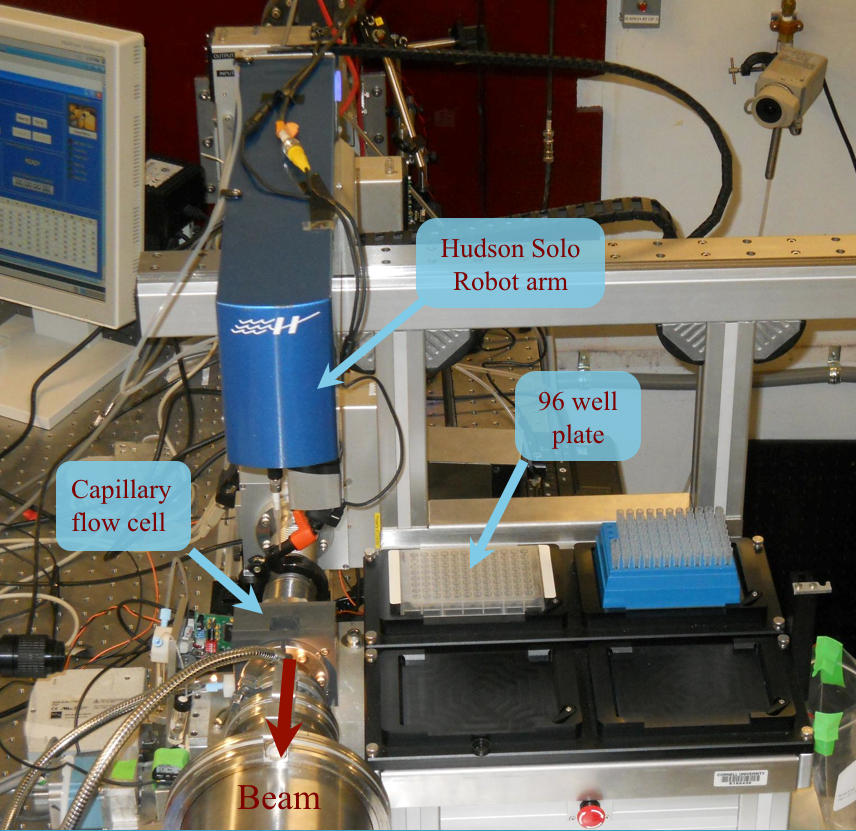
To meet the growing demand for training, MacCHESS offered its first BioSAXS Essentials course. Designed to give students the basic “essentials” necessary to collect and evaluate BioSAXS data, the course offered 1 day of lectures followed by 2 days of practical data collection time. Both F2 and G1 stations were configured for BioSAXS and students were encouraged to bring their own samples. G1 line featured our new BioSAXS high-throughput robot. We limited the first course to 16 students with 9 bringing samples. Due to an overwhelming response, we offered a repeat performance of the course in May that drew 27 students just from the previous course waiting list.

Students were given 3 basic lectures in BioSAXS essential theory interspersed with practical talks from experienced scientists in the field. Software tutorials on data processing and envelope reconstruction where offered Saturday morning. Speakers included Thomas Grant (HWI), Eddie Snell (HWI), Lois Pollack (Cornell), Joseph Curtis (NIST), Nozomi Ando (MIT), Gushol Gupta (UPENN Medical school), Søren Nielsen (MacCHESS), Richard Gillilan (MacCHESS), and Magda Møller (U. Copenhagen). Student groups of 3-4 were allocated 6 hours of beamtime each on the two stations thoughout the weekend.
MacCHESS plans to offer this popular minicourse periodically in the future. Enter your contact information on the official course web page if you want to be added to the notification list for future offerings: www.chess.cornell.edu/BioSAXS course.
Submitted by: Richard Gillilan, MacCHESS, Cornell University
