X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
Human cells have methods to distinguish their own RNA from the RNA of invading viruses. One of those methods involves retinoic acid-inducible gene-I (RIG-I)(also known as DDX58), one of the receptors of the innate immune system in our body: the 2011 Nobel prize is awarded to scientists for their discovery of the innate immune defense systems. RIG-I is a helicase that recognizes and is activated by a number of features which are common to many viral RNAs but not to normal cellular RNA. Once it is activated, it turns on the innate immune response that establishes an anti-viral defense system in the cell. A better understanding of the function of RIG-I could lead to better anti-viral therapies as well as treatments for the consequences when the RIG-I system goes wrong, which can include cancer and auto-immune diseases.
A team of researchers from Rutgers, The State University of New Jersey and UMDNJ-Robert Wood Johnson Medical School, led by principal investigators Joseph Marcotrigiano and Smita Patel, along with Michael Gale Jr. of the University of Washington, have now gained insight into this ability to recognize foreign RNA by solving the crystal structure of the RNA-binding portion of RIG-I together with a piece of double-stranded RNA. The overall structure shows how RIG-I wraps around the RNA, and which amino acid side chains of the protein interact with which features of the RNA. Data for this crystal structure was collected at CHESS beamline F1, APS and NSLS synchrotron facilities.
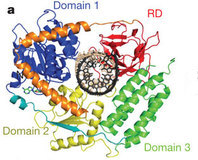
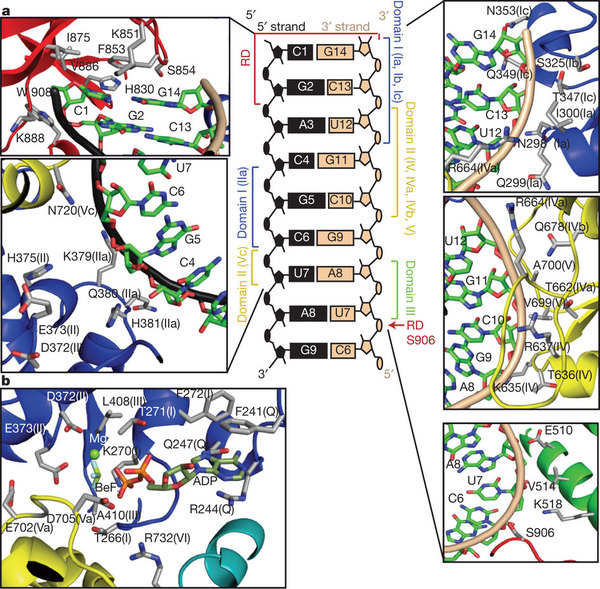
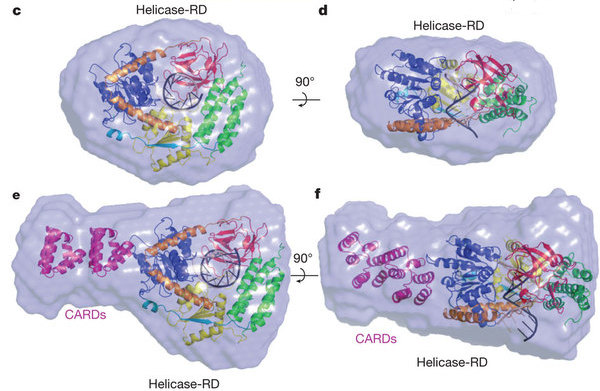
In addition to the crystal structure, the team also used several other techniques to help determine how the RIG-I molecule may change shape when it binds RNA, and how it might activate cellular defenses. One of these techniques, SAXS (Small Angle X-ray Scattering), provides information about the shape of the molecule in solution, rather than in a crystal. The researchers collected SAXS data from CHESS beamline G1 in order to compare the RNA-binding portions of RIG-I revealed in the crystal structure with a more complete molecule of RIG-I, which also contains caspase recruitment domains (CARDs). The structure of the CARDs is known from previous research (PDB entry 2VGQ). CARDs are involved in raising the innate immune response once foreign RNA has been detected.
The research paper can be found in the journal Nature: Fuguo Jiang, Anand Ramanathan, Matthew T. Miller, Guo-Qing Tang, Michael Gale, Smita S. Patel, and Joseph Marcotrigiano; "Structural Basis of RNA Recognition and Activation by Innate Immune Receptor RIG-I", Nature 10537, doi:10.1038 (2011)
The coordinates of the RIG-I crystal structure can be found in Protein Data Bank entry 3TMI.
A press release from the Robert Wood Johnson Medical School contains more details about the researchers and their work.
Another press release can be found from Rutgers.
Submitted by: David J. Schuller, MacCHESS, Cornell University
