X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
Bacteria are constantly bombarded by foreign genetic elements in the form of viruses, plasmids and transposons. Within the last decade it has been discovered that many prokaryotes (one-celled organisms without a nucleus) possess an adaptive immune system for fighting these invaders at the nucleic acid level with guide RNAs. This follows an earlier discovery that some eukaryotes have an analogous anti-sense RNA defense called RNAi (i for interference). It will be important to understand these bacterial defense systems since bacteriophage (viruses which infect bacteria) are used as therapy to treat bacterial infections in humans and animals and as an anti-bacterial measure in meat processing.
This RNA-based bacterial defense system, CRISPR (Clustered regularly interspaced short palindromic repeats) consists of short stretches of variable DNA from pathogens inserted between constant regions of DNA, and a set of genes for CAS (CRISPR-associated) proteins which process the RNA transcribed from CRISPR and form a complex with it to fight off foreign genes. CRISPR is quite widespread, with versions found in about 45% of sequenced bacterial and 83% of sequenced archaea genomes. Among those studying CRISPR systems are Cornell Associate Professor Ailong Ke and his post-doctoral research associate Ki Hyun Nam.
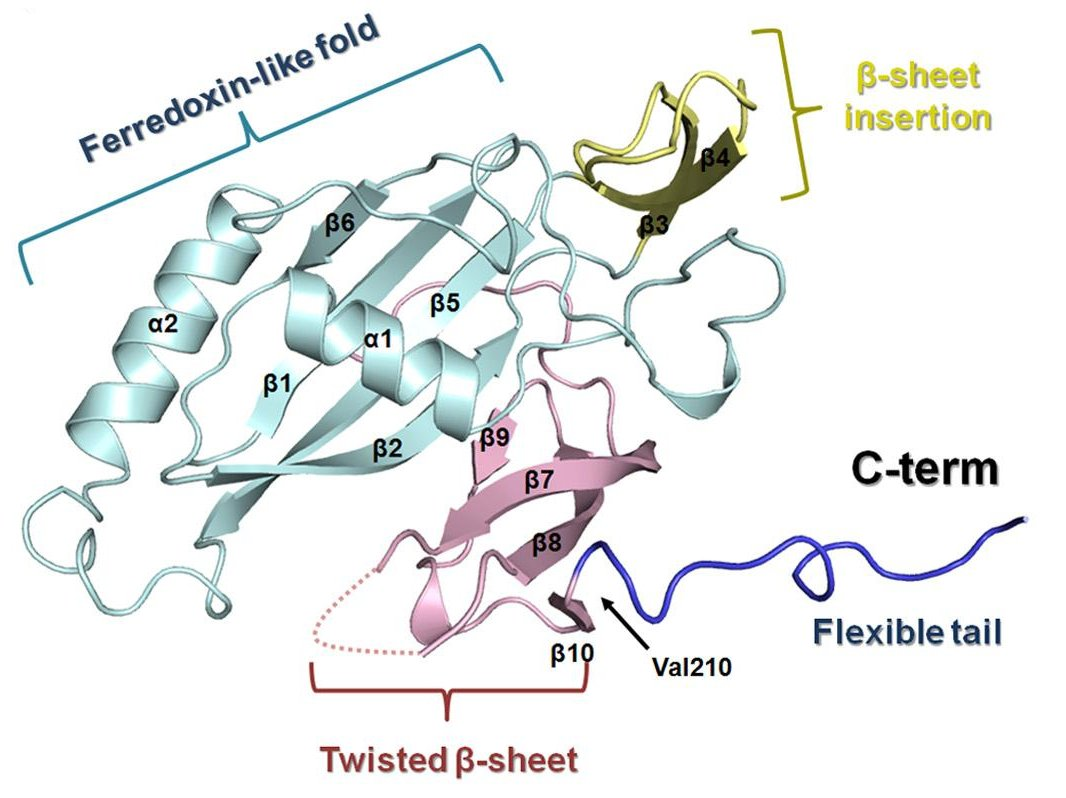
Recently, Nam, Ke and their collaborators explored the Type I-C system from Bacillus halodurans. Through a series of assays, they found that the Cas5d protein is responsible for initiating the CRISPR interference by processing the transcribed CRISPR RNA in a site-specific cleavage reaction, and characterized details of substrate recognition mechanism. Then they crystallized the Cas5d protein and solved its structure with diffraction data collected at CHESS beamline A1. The structure provided details of the RNA binding interface and suggested which residues may be involved in the RNA cleavage. This was further explored with a series of “alanine scanning” experiments to verify the effect of mutating certain residues on the activity of the protein. The X-ray structures were then combined with electron micrographs of the CASCADE (CRISPR-associated complex for antiviral defense) complex to determine how Cas5d might combine with the processed CRISPR RNA and other CAS proteins in a multi-subunit RNA-protein complex to recognize and fight the invading genetic material.
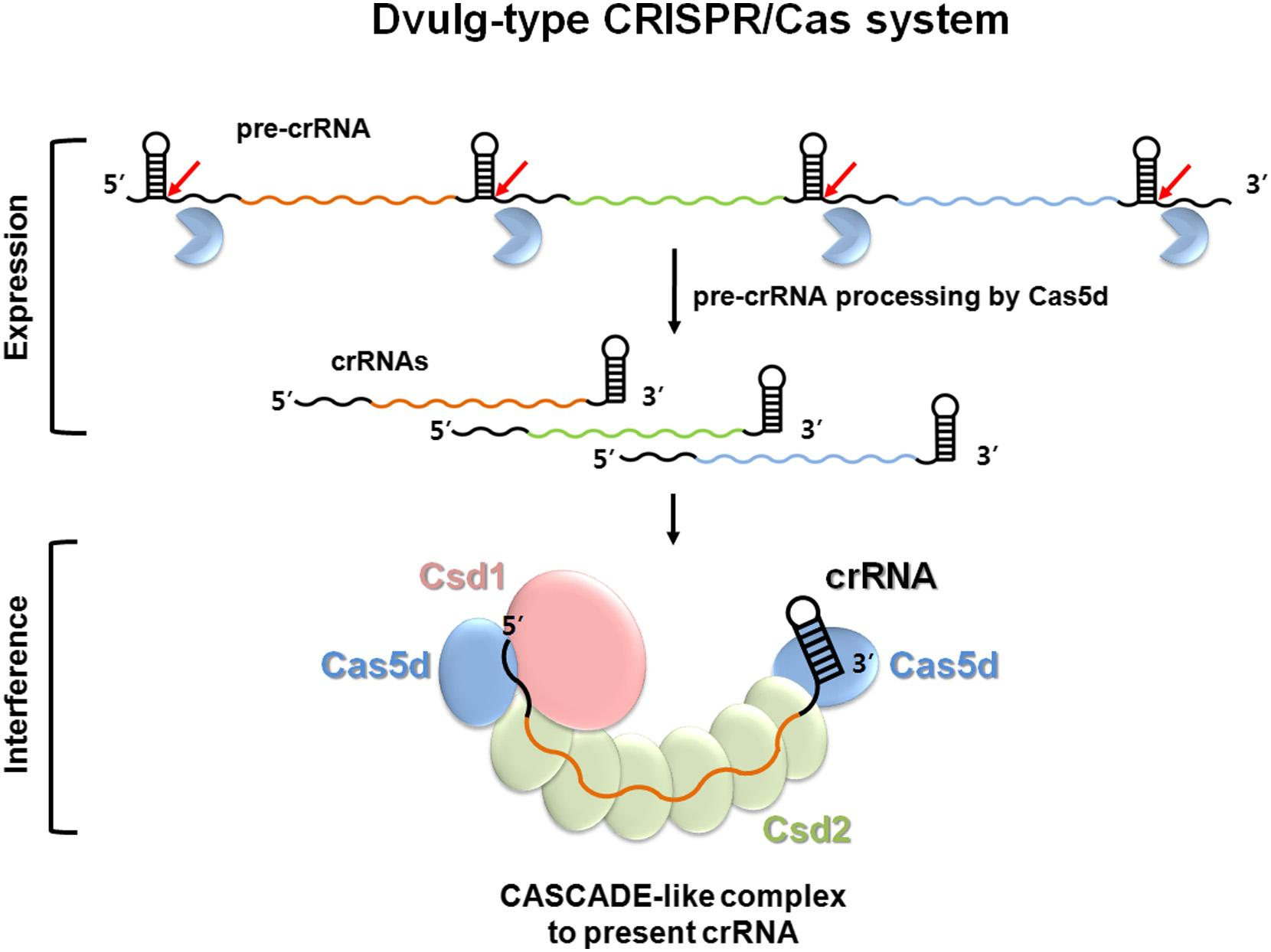
The results of these studies were published recently in the journal Structure: Ki Hyun Nam, Charles Haitjema, Xueqi Liu, Fran Ding, Hongwei Wang, Matthew P. DeLisa, and Ailong Ke; "Cas5d Protein Processes Pre-crRNA and Assembles into a Cascade-like Interference Complex in Subtype I-C/Dvulg CRISPR-Cas System", Structure Volume 20, Issue 9, 5 September 2012, Pages 1574–1584 doi: 10.1016/j.str.2012.06.016 PDB entry: 4F3M.
An accompanying article provides some background and discusses the significance of the work: Nicole A. LeRonde-LeBlanc; "Defense Systems Up: Structure of Subtype I-C/Dvulg CRISPR/Cas", Structure, Volume 20, Issue 9, 5 September 2012, Pages 1574-1584, doi: 10.1016/j.str.2012.08.015.
But this is not all of the CRISPR-related structural biology work Ke and Nam have done. In 2011, they characterized another CAS protein and solved its structure with data from CHESS. The resulting publication highlighted as the Best 2011 JBC papers in the Journal of Biological Chemistry.
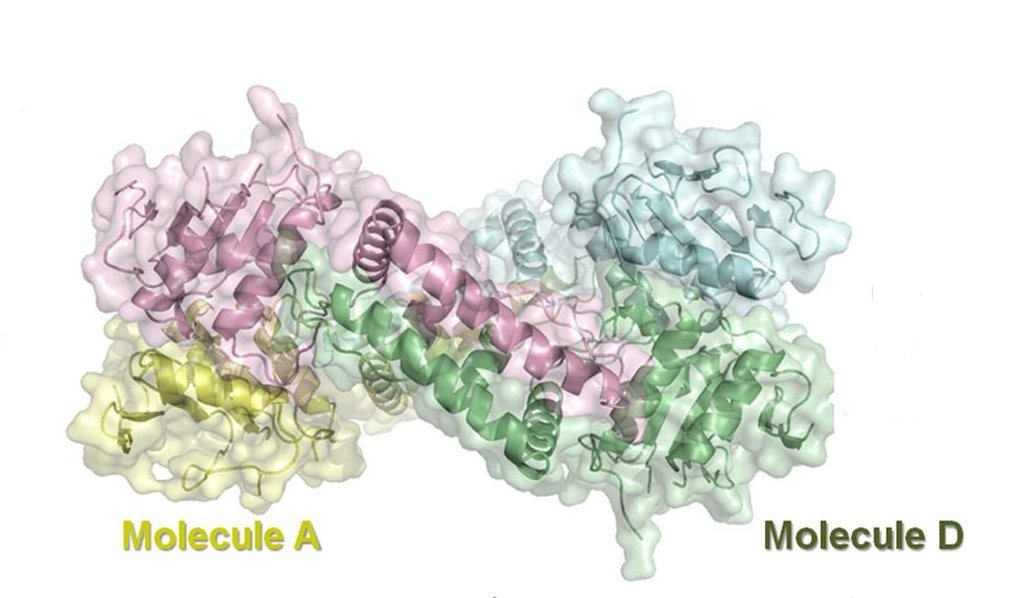 Ki Hyun Nam, Igor Kurinov, and Ailong Ke; "Crystal Structure of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-associated Csn2 Protein Revealed Ca2+-dependent Double-stranded DNA Binding Activity", September 2, 2011 The Journal of Biological Chemistry, 286, 30759-30768.
doi: 10.1074/jbc.M111.256263
PDB entry: 3S5U.
Ki Hyun Nam, Igor Kurinov, and Ailong Ke; "Crystal Structure of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-associated Csn2 Protein Revealed Ca2+-dependent Double-stranded DNA Binding Activity", September 2, 2011 The Journal of Biological Chemistry, 286, 30759-30768.
doi: 10.1074/jbc.M111.256263
PDB entry: 3S5U.
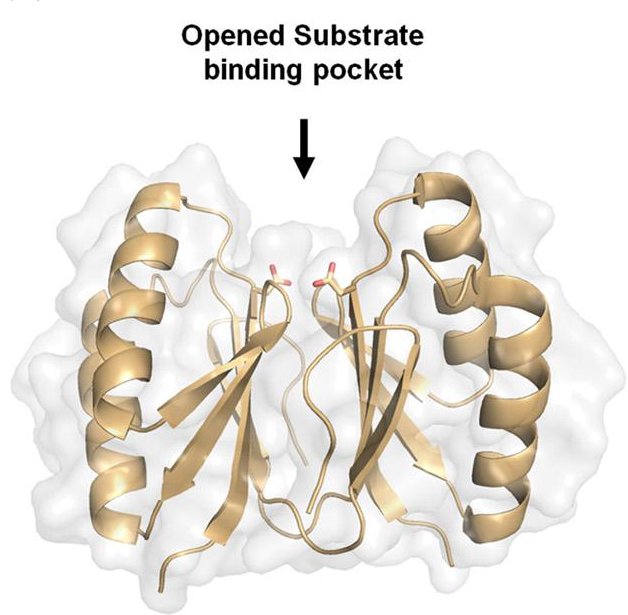 But wait, there's more! Nam and Ke have two more CRISPR-related studies with CHESS X-ray structures on the way soon:
But wait, there's more! Nam and Ke have two more CRISPR-related studies with CHESS X-ray structures on the way soon:
Ki Hyun Nam, Fran Ding, Charles Haitjema, Qingqiu Huang, Matthew P. DeLisa, and Ailong Ke; "Double-stranded Endonuclease Activity in Bacillus halodurans Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-associated Cas2 Protein", October 19, 2012 The Journal of Biological Chemistry, 287, 35943-35952. (Cover Art.) doi: 10.1074/jbc.M112.382598 PDB entries: 4ES1, 4ES2, 4ES3.
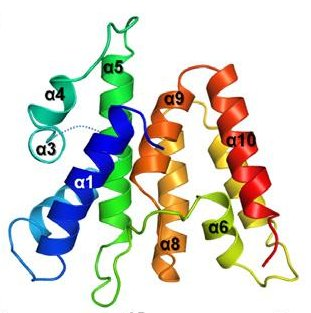 Ki Hyun Nam, Qingqiu Huang, and Ailong Ke; "Nucleic
Acid Binding Surface and Dimer Interface Revealed by
CRISPR-associated CasB Protein Structures", FEBS Letters, in press
PDB entries: 4H79, 4H7A.
Ki Hyun Nam, Qingqiu Huang, and Ailong Ke; "Nucleic
Acid Binding Surface and Dimer Interface Revealed by
CRISPR-associated CasB Protein Structures", FEBS Letters, in press
PDB entries: 4H79, 4H7A.
Qingqiu Huang is a research associate at MacCHESS.
Submitted by: David J. Schuller, MacCHESS, Cornell University
11/01/2012
