X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
Proteins must fluctuate in order to perform cellular functions, such as enzymatic catalysis, protein-protein interactions, and interactions with DNA and RNA. When proteins are cooled the fluctuations dampen and eventually stop, typically at 200 - 240 K. This is called a protein dynamical transition. Proteins below the transition temperature show no appreciable biological function. Above the transition temperature flexibility is restored and the protein becomes increasingly biologically active.
The underlying physical origin of the protein dynamical transition is controversial. Water is thought to be involved, since proteins below the transition temperature behave as though they are dried. But the exact nature of the water-protein coupling is not clearly understood. Does the reduction in temperature dampen fluctuations in the protein, or is the stiffening of the surrounding water primarily responsible? Why does protein dynamical transition occur at around 220 K?
Cornell scientists, using the Cornell High Energy Synchrotron Source (CHESS), have gained new insight into the underlying mechanisms of protein dynamical transition. Their paper, whose first author is MacCHESS staff scientist Chae Un Kim, was published in Proceedings of the National Academy of Sciences [1]. Kim’s co-authors are Mark W. Tate, senior research associate in the Laboratory of Atomics and Solid State physics; and Sol M. Gruner, professor of physics and CHESS director.
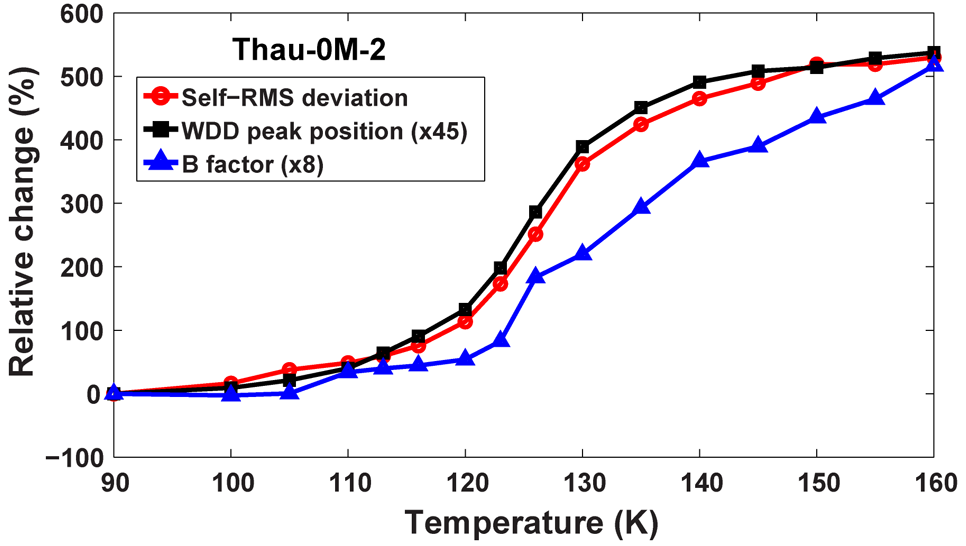
Fig 1: Cryogenic protein dynamical transition around 120 K indicated by the abrupt increase in the Debye–Waller factor (B-factor) and the structural root mean square (RMS) deviation. The water diffuse diffraction (WDD) peak shows the phase transition from high-density (below 110 K) to low-density amorphous (above 140 K) states of water. From [1].
In their experiments, the researchers observed such a transition at 120 K – a much lower temperature – in a protein crystal when cryogenically cooled water confined in the crystal underwent unusual phase transitions (Fig. 1). The protein crystal samples were treated by a high-pressure cryocooling method developed by the researchers [2]. The samples were frozen to liquid nitrogen temperature (77 K) in helium gas pressurized to 2,000 atmospheres. Previously, they had shown this method induces an unusual, high-density amorphous state of water and, upon warming, it transforms to low-density amorphous ice [3, 4].
The protein dynamical transition was then probed by the temperature-controlled X-ray protein crystallography at CHESS to measure the distribution of fluctuation states of a protein. The researchers observed that the fluctuation states suddenly increased as the water underwent the high-density to low-density transition. It appears that the change in the state of the water gave the proteins freedom to fluctuate and wiggle (Fig. 2). This result suggests that the protein dynamical transition was actually enabled by motional freedom provided by the surrounding water.
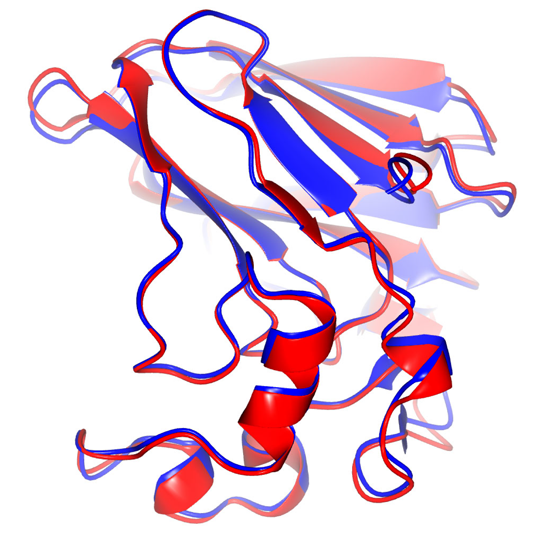
Fig 2: Superposition of Thaumatin structures before (blue, 80 K) and after (red, 160 K) the cryogenic dynamical transition. It demonstrates that the structural displacements are spread out over the whole protein molecule, indicating that the protein dynamical transition is due to global protein dynamical motions. From [1].
In the past, such studies have been experimentally hampered by spontaneous crystallization of water ice. The novel high-pressure method bypasses this problem to allow probing of protein dynamics and the relationship to the phase behavior of water at cryogenic temperatures. The results provide evidence that the physical origin of a protein dynamical transition is driven by water fluctuations, and they also provide insights into the unusual physical properties of supercooled water.
A story on this work may also be found in the Cornell Chronicle:
http://www.news.cornell.edu/stories/Dec11/proteinCHESS.html
or (PDF)
References:
[1] Chae Un Kim, Mark W. Tate,
and Sol M. Gruner; "Protein Dynamical Transition at 110 K", PNAS 108,
20897-20901 (2011)
[2] Chae Un Kim, Raphael Kapfer, and Sol M. Gruner; "High Pressure Cooling of Protein Crystals without Cryoprotectants", Acta Cryst. D61, 881-890 (2005)
[3] Chae Un Kim, Yi-Fan Chen, Mark W. Tate, and Sol M. Gruner; "Pressure Induced High-Density Amorphous Ice in Protein Crystals", J. Appl. Cryst. 41, 1-7 (2008)
[4] Chae Un Kim, Buz Barstow, Mark W. Tate, and Sol M. Gruner; "Evidence for Liquid Water During the High-Density to Low-Density Amorphous Ice Transition", PNAS 106, 4596-4600 (2009)
Submitted by: Chae Un Kim, MacCHESS, Cornell University
01/11/2012
