X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
A group of Rutgers researchers has solved X-ray crystal structures of the RNA polymerase (RNAP)-promoter open complex, a key structure for understanding transcription initiation. Transcription is the copying of a gene to make a complementary RNA, which may then be translated by a ribosome to make a protein. Transcription and translation, along with DNA replication, are the three most fundamental molecular processes within cells. Before transcription the RNA polymerase, aided by initiation factors, must recognize the promoter, a special DNA sequence which marks the start of a gene. While various structures of the RNA polymerase have been solved, either alone or in complexes, these new structures are the first of a specific, promoter-dependent, initiation-factor-dependent transcription initiation complex.
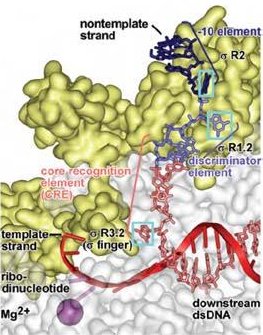
The new structures show in molecular detail how both the RNA polymerase and the accompanying σA transcription initiation factor from the bacterium Thermus thermophilus unwind the DNA double helix and then unstack and flip out bases of the unwound DNA in order to recognize the specific sequence of the promoter and the start of the gene. The unstacked and flipped out bases come from the DNA nontemplate strand, not the one which is copied.
The new structures also show that another part of σA interacts with the DNA template strand, the one that is copied, 'pre-organizing' it to adopt a helical conformation and to engage the RNA polymerase active center.
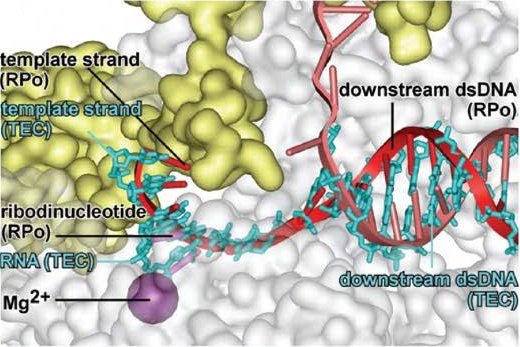
In addition to the RNAP-promoter open complex, the researchers also solved a structure of the complex together with a ribodinucleotide primer, which reveals the interaction of the complex will have with the transcribed RNA once transcription begins. The structures also divulge details about interaction of the polymerase with downstream sections of the DNA, and the conformation of the RNAP clamp within the complex.
The register of the DNA strand within the complex was verified by the substitution of 5BrU (5-bromodeoxyuridine) for a T at a certain position of the DNA sequence (+1 relative to the start site of transcription) in a third structure. The bromine atom provides an 'anomalous scattering' signal to the X-ray diffraction, which makes it possible to precisely locate that atom in crystallographic maps.
X-ray diffraction data used to solve these structures were collected both at CHESS and at the National Synchrotron Light Source.
Protein Data Bank (PDB) entries 4G7O, 4G7Z, 4G7H
Yu Zhang, Yu Feng, Sujy Chatterjee, Steve Tuske, Mary X. Ho, Eddy Arnold, and Richard H. Ebright; "Structural Basis of Transcription Initiation", Scienc Express 18 (October 2012) DOI: 10.1126/science.1227786
Submitted by: David J. Schuller, MacCHESS, Cornell University
11/16/2012
