X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
Synthesis of DNA is an essential foundation for all life on Earth. In all organisms, the precursors for DNA (deoxyribonucleotides) are made from the precursors for RNA (ribonucleotides) using an enzyme called ribonucleotide reductase (RNR). RNRs are classified by the metal-containing cofactor that is used to generate a radical essential for catalysis. Class Ia RNRs (used by all eukaryotes and many aerobic bacteria) are unusual in that the nucleotide-binding sites and the radical-generating metallocofactor are housed in separate homodimeric proteins, called α2 and β2 (Fig. 1). Thus, for each turnover, the two proteins must interact and engage in long-range radical transfer. Furthermore, the nucleotide-binding protein α2 can bind downstream deoxyribonucleotides and ATP in two additional sites other than the active site – these allosteric sites enable RNRs to sense and control the overall levels and balance of intracellular deoxyribonucleotide pools, much like a thermostat that regulates the temperature of a room.
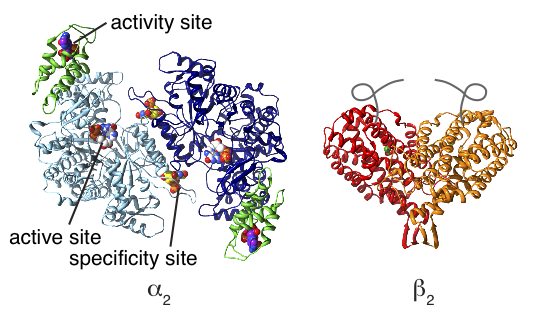
Figure 1 (adapted from Fig. 1 in reference [1])
A major roadblock towards understanding how activity regulation works in class Ia RNRs has been the transient interactions of α2 and β2. To meet this challenge, the group of Catherine Drennan (MIT), in collaboration with JoAnne Stubbe (MIT) and Francisco Asturias (Scripps), investigated the model RNR from E. coli using four complementary techniques: small-angle X-ray scattering (SAXS), analytical ultracentrifugation (AUC), single-particle electron microscopy (EM), and X-ray crystallography.
The team of researchers found that E. coli RNR exists in three major states (Fig. 2), whose distributions are sensitive to nucleotide and protein concentrations. In the presence of the down-regulating allosteric effector, dATP, a large inactive complex dominates. This complex was determined by EM, crystallography, and SAXS to adopt an unusual α4β4 ring structure, composed of alternating α2 and β2 (Fig. 2, right). Residues important for radical transfer from β2 to α2 are separated across the hole of the ring, thus making this complex inactive. Furthermore, the researchers used SAXS to investigate the conformational changes driven by dATP. By applying deconvolution analyses and ab initio shape reconstruction methods to their SAXS data, they were able to determine the mass and shape of the active complex in equilibrium with the dATP-inactivated complex. In the best-fit model for the active complex, α2 and β2 are docked in a globular and compact arrangement that would allow for radical transfer (Fig. 2, middle).
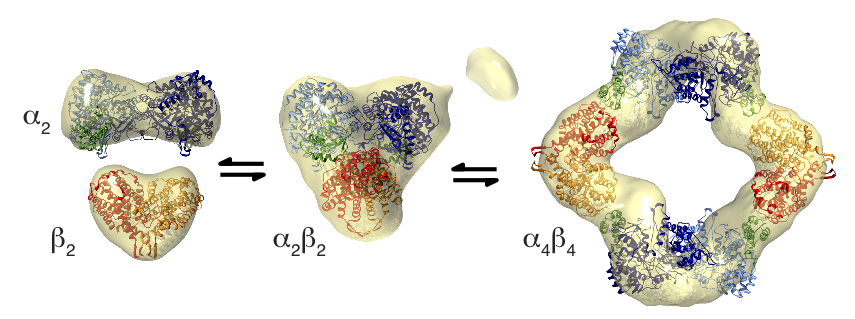
Figure 2 (adapted from Fig. 4 in reference [1])
Solution-based SAXS, performed at the CHESS G1 station, was a key component of this study as E. coli RNR is highly dependent on solution condition, being composed of proteins that are not only weakly interacting but also forming multiple distinct complexes. A number of SAXS-based analyses are presented in this work, including determination of mass, subunit stoichiometery, and shape, as well as the deconvolution of mixtures. The research is reported in PNAS. The first author of the paper, Nozomi Ando, is a post doctoral fellow in the Drennan group (MIT).
Reference:
[1] Nozomi Ando, Ed Brignole, Christina Zimanyi, Michael Funk, Kenichi Yokoyama, Francisco J. Asturias, JoAnne Stubbe,
and Catherine L. Drennan; "Structural Interconversions Modulate Activity of E. coli
Ribonucleotide Reductase", PNAS 108,
21046-21051 (2011)
Submitted by: Chae Un Kim, MacCHESS, Cornell University
01/06/2012
