X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
Influenza is a severe infectious disease, caused by RNA viruses, that can spread around the world in seasonal epidemics. Moreover, influenza can become pandemic when a new strain of the virus appears in humans, as has happened several time in the 20-th century (1918 H1N1 - Spanish flu, 1957 H2N2 - Asian flu, 1968 H3N2 - Hong Kong flu, 2009 H1N1 - swine flu). The primary defense against influenza is vaccination in the form of either inactivated or live attenuated viruses. However, in the event of a pandemic, timely production of vaccines, together with their partial efficacy, present serious challenges to the world's health institutions. The recent outbreak of an unusual H7N9 strain, the emergence of drug-resistant variants of influenza A strain, and the adaptation of H5N1 for human-to-human transmission emphasize the urgent need for developing new broadly effective flu drugs.
A group of researchers from Rutgers University led by Dr. E. Arnold has discovered and developed a new class of inhibitors targeting influenza endonuclease activity of the viral polymerase. Previous studies have determined that the endonuclease active site resides in the N-terminal domain of the polymerase acidic protein subunit (PAN), and crystal structures of PAN have revealed a deep active site cleft containing multiple subpockets that can be targeted for drug design (Figure 1.)
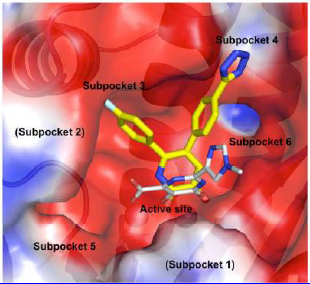
Figure 1. Calculated electrostatic potential surface of the active site cleft of PAN, with different subpockets labeled. Binding of two different inhibitors is shown: a carboximide derivative with 3 chelating groups [1] (gray) and one of the group studied in the new work (yellow).
Highly diffracting crystals (1.7 Å) of constructs engineered from pandemic 2009 H1N1 influenza were obtained and were soaked in solutions containing 10 mM small molecule fragments. Fragment screening of a diverse library of inhibitors by X-ray crystallography identified a new class of PAN - inhibiting compounds chelated to the two catalytic metal ions, M1 and M2, in a mode similar to the known endonuclease inhibitors. In addition to M1 and M2, the presence of a third metal ion (M3) in the active site cleft was discovered and subsequently verified by soaking of high concentrations of various metals into crystals of PAN. X-Ray crystallography experiments were largely conducted at CHESS F1 beamline and NSLS at Brookhaven National Laboratory. High-resolution crystallographic data confirmed the binding of a metal ion in the M3 site in the presence of an inhibitor (compound 1 in Figure 2A); this observation is potentially relevant to the mechanism of endonuclease activity (Figure 2).
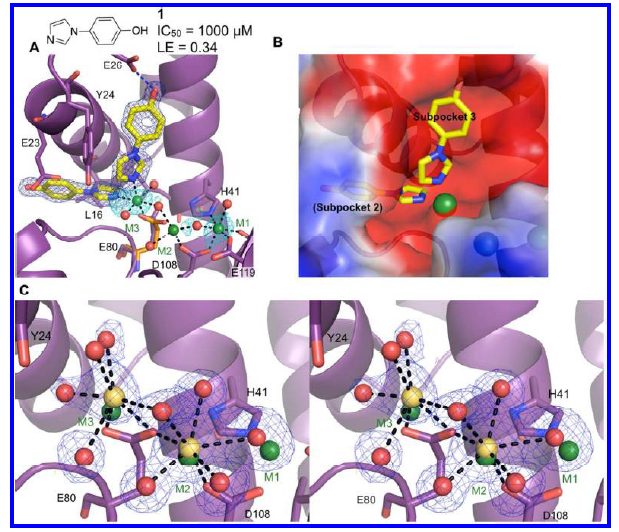
Figure 2. Discovery of a third active site metal. Metal-coordinating bonds are depicted as black dashed lines and hydrogen bonds as blue dashed lines. Residues with significant structural changes upon binding of a ligand are displayed with the apo structure shown in orange. (A) Structure of 1 bound to subpockets 2 and 3. Compound 1 chelates a third metal (M3), which coordinates Glu80. The electron density (dark blue mesh) is calculated from a Fo-Fc omit map and contoured at 5σ. Electron density calculated from anomalous data (cyan) is contoured at 3σ. (B) Structure of 1 with calculated electrostatic potential surface of active site and subpocket labels. (C) Stereoview of the active site cleft, showing electron density for the metal ions (Ca2+, yellow) and coordinating waters (red) in the structure of apo PAN soaked in 100 mM CaCl2. For comparison, the metal ions from the structure with 1 are shown in green. This confirms that M3 is not an artifact of binding compound 1.
The most active of the new class of inhibitors discovered by fragment screening (followed by multiple rounds of optimization using enzyme assays, crystal structures, molecular modeling, and medicinal chemistry), has significant antiviral activity (EC50 11 μM) and represents a potential treatment for flu.
The manuscript [2] has just been accepted for publication in journal of ACS Chemical Biology.
http://pubs.acs.org/doi/full/10.1021/cb400400j
References:
- Dubois, R.M., Slavish, P.J., Baughman, B.M., Yun, M.K., Bao, J., Webby, R.J., Webb, T.R., White S.W. (2012), “Structural and biochemical basis for development of influenza virus inhibitors targeting the PA endonuclease”. PloS Pathog. 8, No. E10002831.
- Bauman, J.D., Patel, D., Baker, S.F., Vijayan, R. S. K., Xiang, A., Parhi, A.K., Martinez-Sobrido, L., LaVoie, E.J., Das, K., Arnold, E. (2013), “Crystallographic fragment screening and structure-based optimization yields a new class of influenza endonuclease inhibitors”, ACS Chem. Biol. 2013 Sept. 13 [Epub ahead of print].
Submitted by: Irina Kriksunov, MacCHESS, Cornell University
10/10/2013
