X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
Eukaryotic cells, from ‘complex’ multicellular organisms to ‘simple’ single-celled microorganisms, possess an extensive network of membrane-bound organelles that are necessary to carry out a variety of processes essential for life, such as metabolism and extensive cell signaling pathways. Cells have developed an elaborate trafficking pathway to maintain both the protein and lipid compositions of organelles. Central to the trafficking pathway is the trans-Golgi network (TGN), an organelle responsible for modifying and sorting a vast number of proteins destined for organelles. In order to accomplish this sorting, cells employ a number of cytosolic proteins (proteins not integrated into the membranes of organelles) that bind to both lipid and protein alike to properly sort and shuttle the right components at the right time. Of particular interest is the trafficking of proteins from the TGN to the exterior of the cell or plasma membrane (PM). In this trafficking step, proteins are sorted into small membrane-bound organelles called secretory vesicles that are brought to the surface of the cell by an extensive filament network. However, it is not completely understood how proteins are sorted into these vesicles.
Researchers have studied the budding yeast (Sacchromyces cerevisae) and an enzyme called chitin synthase III or Chs3 (a protein responsible for synthesizing chitin for the yeast cell wall), and have gained insights into this particular step of trafficking. It was discovered that this particular protein gets shuttled back and forth between the TGN and PM in a tightly regulated manner (Chuang and Schekman, 1996). Subsequently, it was shown that this localization within the cell was dependent upon a set of cytosolic proteins, Chs5, Chs6, Bud7, Bch1, and Bch2, which was named as the exomer complex (Trautwein et al., 2006). The exomer is referred to as a cargo adaptor, for its role of sorting cargo, such as Chs3, into vesicles. Previous biochemical analysis of the complex revealed that the core exomer protein, Chs5, could interchangeably interact with any of the four other proteins to form the complex (Wang et al., 2006). Now, the Fromme group at Cornell University solved the crystal structure of a tetrameric exomer complex at 2.75 Å resolution and the results are reported in EMBO Journal (Paczkowski et al., 2012). The crystallographic data set was collected at the CHESS A1 beamline and the structure was determined using multiple isomorphous replacement with anomalous scattering (MIRAS).
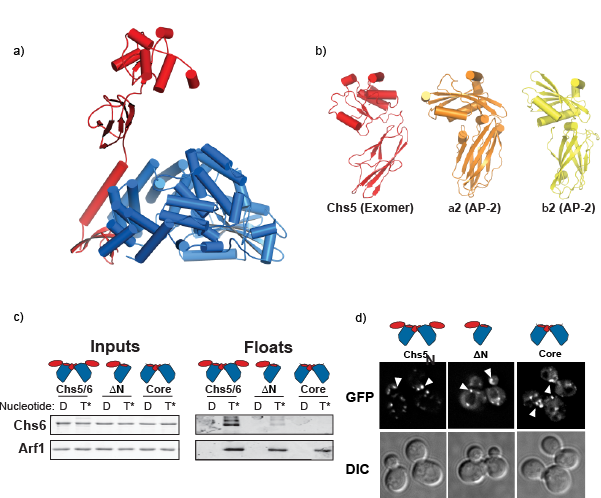
Figure 1. a) The crystal structure of Chs5 (red) and Chs6 (blue) from the exomer complex. b) Structural resemblance of the Chs5 FBE domain (this work) and the appendage domains of other cargo adaptors (PDB: 1B9K & PDB: 1E42) (Owen et al., 1999 and Owen et al., 2000). c) The Chs5 FBE domain is required for exomer interaction with Arf1. d) Multiple domains in Chs5 are required for complete localization to the TGN. Internal puncta within the cells correspond to the TGN.
The structure provides insights into how the individual proteins, Chs5 and Chs6, of the complex interact through a long α-helix (figure 1a). Additionally, the structure reveals an interesting flexible domain on Chs5, known as the FBE domain, which extended away from the bulk of the complex (figure 1a) and is required for exomer function. Interestingly, this domain is topologically similar to domains found in other cargo adaptors despite the lack of sequence homology (figure 1b). Previously, it had been shown that exomer is recruited to membranes through a direct interaction with Arf1 (a well-conserved eukaryotic protein that is involved in many trafficking steps) as well by interacting with the membranes themselves (Trautwein et al., 2006 & Wang et al., 2006). Using a biochemical approach, the Fromme group shows that the novel FBE domain interacts with Arf1, and that the FBE domain is required for recruitment of the complex to the membrane surface (figure 1c), while Chs6 is responsible for Arf1-independent membrane association. Further, by making truncations to the Chs5 protein, its localization can be analyzed in a living cell to determine the requirements for associating with the TGN in vivo (figure 1d). The findings indicate that multiple domains cooperate to bring exomer to the TGN, and that exomer’s localization to the TGN is dependent upon its interactions with the membrane, its cargo, and the universal trafficking regulator, Arf1.
See also Sorting proteins to the right address at the cellular post office for additional related work by the Fromme group.
References:
Chuang JS, Schekman RW (1996) Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p.
J Cell Biol 135: 597–610
Trautwein M, Schindler C, Gauss R, Dengjel J, Hartmann E, Spang A (2006) Arf1p, Chs5p and the ChAPs are required for export of specialized cargo from the Golgi.
EMBO J 25: 943–954
Wang CW, Hamamoto S, Orci L, Schekman R (2006) Exomer: A coat complex for transport of select membrane proteins from the trans-Golgi network to the plasma membrane in yeast.
J Cell Biol 174: 973–983
Paczkowski JE, Richardson BC, Strassner AM, Fromme JC (2012) The exomer cargo adaptor structure reveals a novel GTPase-binding domain.
EMBO J. 31: 4191-4203
Owen DJ, Vallis Y, Noble ME, Hunter JB, Dafforn TR, Evans PR, McMahon HT (1999)A structural explanation for the binding of multiple ligands by the alpha-adaptin appendage domain.
Cell 97: 805–815
Owen DJ, Vallis Y, Pearse BM, McMahon HT, Evans PR (2000) The structure and function of the beta 2-adaptin appendage domain.
EMBO J 19: 4216–4227
Submitted by: 5/24/2013
