X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
Ribonucleotide reductase (RNR) is essential for DNA synthesis and cell proliferation. RNR is an enzyme that catalyzes the conversion of nucleotide diphosphates (NDPs) to deoxynucleotide diphosphates (dNDPs), the building blocks of DNA. RNR plays a critical role in regulating the total rate of DNA synthesis, which is necessary to maintain the constant DNA to cell mass ratio during cell division and DNA repair. The reaction is strictly conserved in all living organisms and proceeds via a free radical mechanism of action. Class Ia RNR (e. g. Escherichia coli RNR) enzymes are constructed from large RNR α2 and small RNR β2 subunits which associate to form an active heterodimeric tetramer: an α2β2 complex. The structure of the active α2β2 complex analog and its intermediate status between dissociated and inhibited states was confirmed by Biological Small-angle X-ray scattering (BioSAXS) experiments conducted at CHESS by MIT researchers from Dr. Drennan's group. In this PNAS article Minnihan et. al. also report an important result on the reaction mechanism in a model predicted by Uhlin and Eklund nearly 2 decades ago.
Nucleotide reduction is initiated when a diferric-tyrosyl radical (Y122•) cofactor in the β2 subunit transiently oxidizes a cysteine in the active site of the α2 subunit. Each turnover requires reversible long-range proton-coupled electron transfer over 35 Å between the two subunits by a specific pathway: Y122• ↔ [W48?] ↔ Y356 in β2 to Y731 ↔ Y730 ↔ C439 in α2 (1). The 35 Å distance between the diferric-Y122• cofactor and the active site cysteine (C439) was predicted in Uhlin and Eklund’s docking model for the active E. coli α2β2 complex, which was based on shape complementarity between the structures of the individual subunits (2). The detailed mechanism for proton-coupled electron transfer is shown in Fig. 1A.
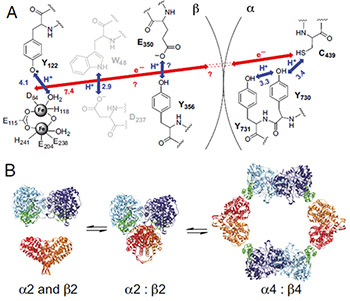
Fig. 1. (A) Model for long-range (∼35 Å), reversible radical transfer in E. coli class Ia RNR. Orthogonal proton-coupled electron transfer (PCET) is proposed to be operative in the β2 subunit, whereas colinear PCET is proposed within the α2 subunit. Y356 has not been observed in any crystal structure of β2; thus, its location relative to the other residues is unknown. W48 and D237 are shown in gray because their participation has not been established experimentally. (B) Three-state model for E. coli class Ia solution equilibrium between free α2 and β2, α2β2, and α4β4.
The scientists from the Drennan lab previously characterized the equilibrium, in solution, across the three inter-converting subunit states: α2 + β2 ↔ α2β2 ↔ α4β4 (Figure 1B, Ref. 3). Now, the group has used several types of experiments to provide evidence for the utilization of an amino acid pathway in long range radical transfer (RT). For better understanding of the RT mechanism, a tyrosine (Y) residue was replaced with the unnatural Y analog (NH2Y) with modified redox properties. The incorporation of NH2Y, especially at position 730 of α2, generated a chemically and kinetically competent NH2Y730• intermediate in the α2 subunit in the presence of wild type (wt)-β2, substrate, and effector. This long-lived radical intermediate proved to be capable of making dNDPs, as well as inducing formation of a kinetically stable α2β2 complex of E. coli RNR class Ia.
Competitive inhibition and affinity chromatography pull-down assays showed that Y730NH2Y-α2 and wt-β2 interact cooperatively and with increased subunit affinity (Kd = 7 nM) compared to the non-cooperative weak interaction(s) between the wt subunits (Kd = 0.18 μM). By conducting stopped-flow (SF) fluorescence experiments, the researchers established that the dissociation rate constant for the Y730NH2Y-α2|wt-β2 interaction is slowed by ∼104-fold relative to wt-α2|wt-β2 (∼60 s−1). Further characterization of the tightly interacting Y730NH2Y-α2 and wt-β2 subunits by single-particle EM revealed an α2β2 complex resembling the original docking model. The structure of Y730NH2Y-α2|wt-β2 complex and its quaternary equilibria were characterized by Biological Small-angle X-ray scattering (BioSAXS) experiments.
BioSAXS experiments were very important in establishing that the α2β2 complex is an intermediate between the subunit dissociated state and the inhibited α4β4 state. A shift in equilibrium toward α4β4 occurs in the presence of increasing protein or inhibitor concentrations, and this form was predominant in the wt case. In the modified protein the enhanced stability of the Y730NH2Y-α2|wt-β2 interaction leads to increased abundance of α2β2 relative to the other states, i. e. the presence of an NH2Y• shifts the subunit equilibrium strongly in favor of a globular α2β2 structure. Data were collected using a Pilatus 100-K detector and an in vacuo 2-mm pathlength quartz capillary flow cell at the G1 station. Results from BioSAXS experiments, revealing the Y730NH2Y-α2|wt-β2 complex stability, are presented in Fig. 2. In addition, activity assays, performed subsequently, indicated that the Y730NH2Y-α2|wt-β2 complex induced by NH2Y730• formation is active or exists in rapid equilibrium with an active state.
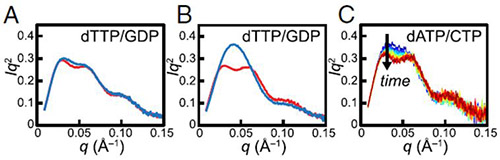
Fig. 2. SAXS reveals the stability of the Y730NH2Y-α2|wt-β2 complex. (A) Pre-turnover mixture of 30 μM wt-α2 with met-β2 (red) and post-turnover mixture of 30 μM wt-α2 with wt-β2 (blue) in the presence of 0.1 mM dTTP and 1 mM GDP display a bimodal Kratky curve, indicative of the presence of α4β4. (B) Pre-turnover mixture of 30 μM Y730NH2Y-α2 with met-β2 in the presence of 0.1 mM dTTP and 1 mM GDP (red) also exhibits a bimodal Kratky curve. Initiation of turnover with Y730NH2Y-α2 and wt-β2 leads to a dramatically different Kratky curve (blue) that is consistent with a stable α2β2 complex. (C) Initiating turnover of 30 μM Y730NH2Y-α2 and wt-β2 in the presence of 175 μM dATP and 1 mM CDP leads to a slow change from a largely monomodal (blue) to bimodal (red) Kratky curve, consistent with the slow conversion of α2β2 to α4β4. Time proceeds from 2–22 min in increments of 2 min.
In conclusion, strong evidence that the reaction of Y730NH2Y-α2, wt-β2, substrate, and effector generates a kinetically stable α2β2 complex, and that the stability of this complex originates from the formation of an NH2Y730 radical on the RT pathway, is presented in a PNAS publication by Minnihan et. al. This work reveals a previously unrecognized regulatory function of the E. coli RNR class Ia: generation of a transiently stabilized α2β2 complex prevents the loss of the catalytic radical during turnover.
Complete article here:
http://www.pnas.org/content/early/2013/02/14/1220691110.short
References:
[1] Stubbe J, van Der Donk WA (1998) "Protein radicals in enzyme catalysis," Chem Rev 98(2):705–762.
[2] Uhlin U, Eklund H (1994) "Structure of ribonucleotide reductase protein R1," Nature 370(6490):533–539.
[3] Ando N et al. (2011) "Structural interconversions modulate activity of Escherichia coli ribonucleotide reductase," PNAS 108(52):21046-21051.
Submitted by: Irina Kriksunov, MacCHESS, Cornell University
01/10/2014
