X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
Within the past decade it was discovered that many cells have an adaptive immune system to fight off foreign RNA or DNA which may have been inserted by a viral, plasmid or transposon attack. In bacteria, this system is known as CRISPR (Clustered Regularly Interspaced Palindromic Repeats, see CHESS news item: Adaptive Immune Systems of Bacteria, 2012) and consists of RNA which recognizes and binds the foreign DNA or RNA through an RNA-mediated interference mechanism and a number of Cas (CRISPR-associated) proteins which help to either form the complex or to degrade the foreign DNA. The complex is known as CASCADE (CRISPR-Associated Complex for Antiviral Defense).
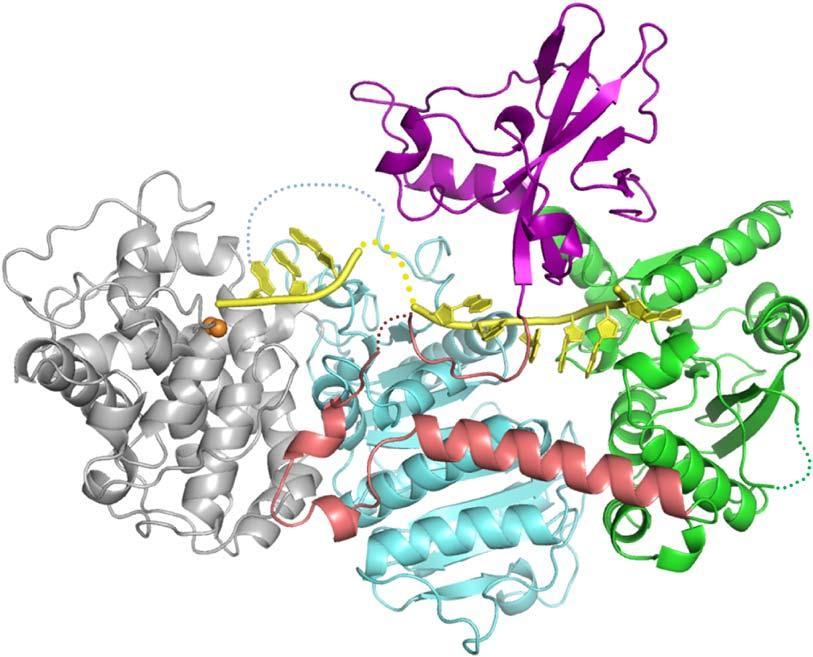
Figure 1. DNA in yellow, iron atoms in orange, proteins domains in various colors.
There are a number of variations of the CRISPR system in assorted types of bacteria; the current work concerns CRISPR type I. This type includes the Cas3 protein which serves both as a helicase to unwind DNA and a nuclease to chop it up. A couple of structures of Cas3 have been solved previously, but the current study by the Ailong Ke laboratory of Cornell University and an international team of collaborators is more complete in that the crystallographically resolved portion includes both the nuclease and helicase domains of Cas3, as well as a 12 nucleotide piece of single-stranded DNA which inhabits the substrate site, and a pair of metal atoms (identified as iron in the current structure by the XAFS technique) which may serve to effect the cleavage of the target DNA.
Thus, the current structure provides new information about how the Cas3 protein is recognized by the CASCADE complex, how the nuclease preferentially receives single-stranded DNA from the helicase, and how the degradation of DNA proceeds. The Ke group solved a number of variant structures with different forms of adenosine nucleotide (ATP, ADP, AMP-PNP) to help resolve the reaction cycle of the helicase. They also performed a number of site-directed mutation studies to glean more information about important residues. The Ke group collected diffraction data on their crystals both at CHESS beamline A1 and at APS NECAT beamline 24-ID-C.
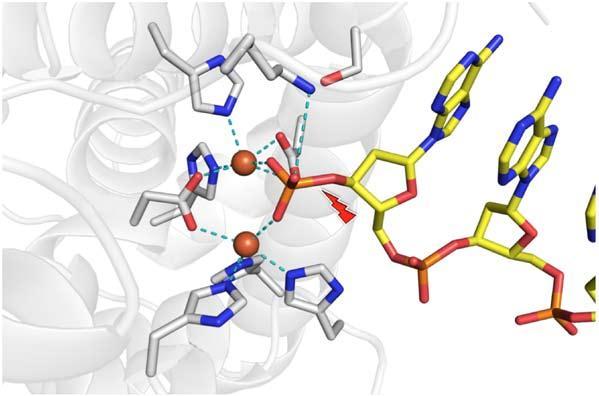
Figure 2. The two metal atoms in the nuclease active site.
Detailed information about bacterial defense systems may have considerable medical and agricultural importance. Bacteriophages and plasmids have potential for use in bacterial control, and overcoming the defense systems bacteria use to fight them off would make the control agents more effective.
Reference:
[1] Yanwu Huo, Ki Hyun Nam, Fang Ding, Heejin Lee, Lijie Wu, Yibei Xiao, M Daniel Farchione Jr, Sharleen Zhou, Kanagalaghatta Rajashankar, Igor Kurinov, Rongguang Zhang, Ailong Ke, "Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation", Nature Structural & Molecular Biology 21, 771-777 (2014).
Submitted by: David Schuller, MacCHESS, Cornell University
09/09/2014
