X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
DNA in the cell must be stored in a compact form (or it wouldn't fit) that also allows it to be translated to RNA, and to be copied when a cell divides. At the lowest level, compaction is achieved by wrapping stretches of DNA around a core of histone proteins to form nucleosomes in a “beads on a string” configuration: 147 base pairs of DNA per nucleosome, with linkers of variable length between nucleosomes. The nucleosome core particle (NCP), or one “bead”, consists of 4 types of histone (H2A, H2B, H3, and H4), each of which has a compact core plus a positively-charged, flexible tail that protrudes from the core. DNA must be made accessible to cellular machinery for it to be processed, which requires at least partial unwrapping from the NCP. The histone tails are clearly involved in control of the unwrapping, but many questions remain about details of the process.
The Andresen group (Gettysburg College) has contributed to answering these questions using a combination of small-angle X-ray scattering (SAXS, a measure of global structure) and fluorescence resonant energy transfer (FRET, a measure of the distance between localized structural features) to study wild-type and modified NCPs. SAXS measurements were performed at CHESS G1, using 30 μL samples in a capillary cell, oscillated across the beam to avoid radiation damage. Data were processed using the RAW software available from MacCHESS. Theoretical scattering curves for comparison with observed data were calculated using the ATSAS package, particularly the program CRYSOL. Comparison of observed and predicted curves (Figure 1) revealed that the depth of the dip in the scattering curve at q=0.14 Å-1 correlates well with the degree of DNA unwrapping.
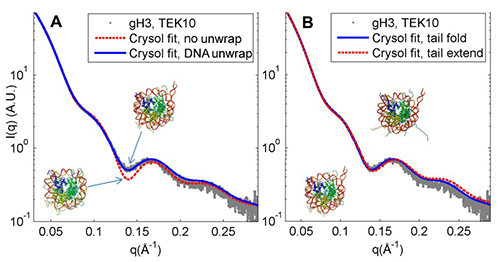
Figure 1. Comparison of observed (gray dots) and predicted (dotted red and solid blue lines) SAXS profiles for NCPs with H3 tail truncated, in a solution with 10 mM KCl. A, model with 20 bp of DNA unwrapped from one end agrees best with observation. B, predictions from models with tail (remaining after truncation) folded and tail extended are nearly identical, so tail extension will not interfere with using SAXS to measure DNA unwrapping.
Using this measure, it was found that truncation of the H3 tail (gH3 form) increases the amount of unwrapped DNA from about 10 (in wild-type, WT) to about 25, while truncation of the H4 tail (gH4 form) does not. The amount of unwrapping is affected by the salt concentration, and at high salt concentration the DNA in gH4 appears to actually be less unwrapped than in WT. FRET measurements of the distance between the two ends of the DNA support more unwrapping in gH3 and less in gH4, relative to WT. Careful analysis of the SAXS data indicates that discrete NCP states with about 10 bp DNA unwrapped and 20 bp DNA unwrapped exist, and that different conditions lead to different fractions of the two states being present. This work contributes to understanding the role of nucleosomes in DNA processing in the cell.
Reference:
[1] K. Andresen, I. Jimenez-Useche, S.C. Howell, C. Yuan, X. Qiu, “Solution scattering and FRET studies on nucleosomes reveal DNA unwrapping effects of H3 and H4 tail removal,” PloS One 8, e78587 (2013).
Submitted by: Marian Szebenyi, MacCHESS, Cornell University
08/13/2014
