X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
RNA polymerase (RNAP) assembles an RNA strand corresponding to the DNA sequence of a gene, in a precisely choreographed series of molecular motions. To understand this critical cellular process, biologists are studying the fine details of the structural changes involved, and how they are regulated. RNAP complexes vary from one species to another, but a core subset of proteins is found throughout archaeal and eukaryotic life forms. Comparison of archaeal and eukaryotic proteins reveals how structural motifs have been modified during evolution, so that function is maintained while regulation has become more complex in eukaryotic species.
The Murakami group (Penn. State) determined the crystal structure of RNAP from the archaeal species Thermococcus kodakarensis, which is believed to be similar to the common ancestor of Archaea and Eukaryota. Diffraction data were collected at CHESS F1 station, and the structure was solved, to 3.5 Å resolution, with the help of known RNAP and RNAP subunit structures. The final refinement gave Rwork/Rfree of 27.7/31.6%.
The overall T. kodakarensis RNAP structure is similar to that of the yeast RNAP Pol II, but has the “clamp” (protein 1 in the figure) in an opened position and the “stalk” (proteins 4 and 7) rotated 12° relative to the Pol II position. The new structure leads to a model for the domain motions involved as the RNAP “clamp” closes about a DNA strand. Concerted motion of the clamp and stalk resolve the problem of steric clashes observed in open models constructed from the Pol II crystal structure by moving only the clamp domain. Alignment of the two structures also helps to identify the surface regions involved in regulation by eukaryotic-specific transcription factors.
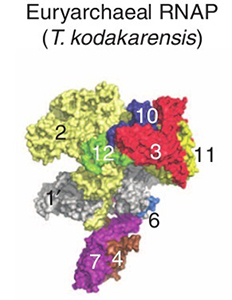
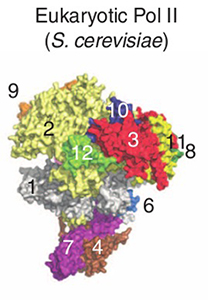
Structures of archaeal and eukaryotic forms of RNAP. Numbers identify individual proteins in the complex. Proteins 4 and 5 are hidden on the far side of the complex; no equivalents to proteins 8 and 9 exist in the T. kodakarensis structure.
References:
[1] S.-H. Jun, A. Hirata, T. Kanai, T. J. Santangelo, T. Imanaka, and K. S. Murakami, "The X-ray crystal structure of the euryarchaeal RNA polymerase in an open-clamp configuration," Nature Commun. DOI: 10.1038/ncomms6132 (2014).
Submitted by: Marian Szebenyi, MacCHESS, Cornell University
11/17/2014
