X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
For the last decade, the Tristram-Nagle/Nagle lab (Carnegie Mellon University) has been working on the interaction of HIV-1 peptides with specific membranes in the HIV virion, T-cell plasma and nuclear membranes (see references below). The peptides are derived from larger proteins that are active in infection, nuclear translocation and budding of new viruses. The current project, peptide MA31, is derived from the N-terminus of the Gag Matrix protein; this has been one focus of three of our trips to CHESS, the most recent in March, 2015.
Their method of measuring diffuse X-ray scattering from fully hydrated membranes allows determination of not only the membrane structure, but also its elastic properties, including the bending modulus, KC. They determined that MA31 lowers the energy required to bend a membrane, especially when a unique negatively charged lipid, PIP2, is present. The effect was smaller for membranes containing another negatively charged lipid, PS, suggesting that there is a specific interaction between PIP2 in the T-cell plasma membrane and the highly basic region of MA31.
They have also determined that MA31 containing a myristoyl group (C14 carbon chain), enters further into the bilayer than does the nonmyristoylated form of MA31. Preliminary electron density profiles are shown below for a single concentration of lipid:peptide (50:1). New samples explored many concentrations of MA31 with/without myristoylation in PIP2 and PS containing membranes. This tendency for the N-terminus of MA31 to locate deeply into the bilayer interior when it is myristoylated, may have implications for targeting the entire MA protein to the plasma membrane prior to virion budding. The MA protein is thought to organize the assembly of the other components of the virus prior to packaging.
This group also collaborates with molecular dynamics simulators; the comparison of X-ray and simulation data is made at the level of the form factors, which are model-independent. The combination of MD simulations with our X-ray diffuse data is a powerful method to precisely determine peptide/lipid membrane structure.
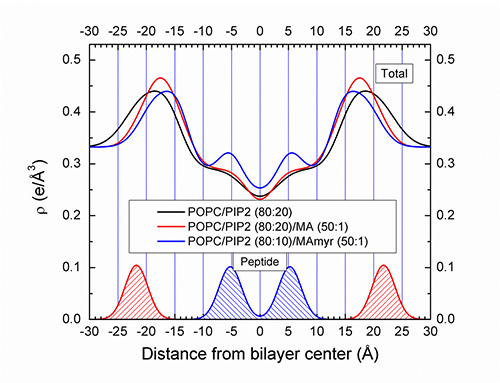
Figure: Preliminary electron density profiles are shown for concentrations of lipid:peptide indicated.
(This work acknowledges R0144976 from the NIGMS at NIH.)
Akabori, K., Huang, K., Treece, B.W., Jablin, M.S., Maranville, B., Woll, A., Nagle, J.F., Garcia, A.E., Tristram-Nagle, S. 2014. HIV-1 Tat membrane interactions probed using X-ray and neutron scattering, CD spectroscopy and MD simulations. Biochimica et Biophysica Acta 1838:3078-3087.
Boscia, A., Akabori, K., Benamram, Z., Michel, J.A., Jablin, M.S., Steckbeck, J.D., Montelaro, R.C., Nagle, J.F. and Tristram-Nagle, S. 2013. Membrane Structure Correlates to Function of LLP2 on the Cytoplasmic Tail of HIV-1 GP41 Protein. Biophysical Journal 105:657-555.
Shchelokovskyy, P., Tristram-Nagle, S. and Dimova, R. 2011. Effect of the HIV-1 Fusion Peptide on the Mechanical Properties and Leaflet Coupling of Lipid Bilayers. New Journal of Physics 13: 025004.
Tristram-Nagle, S., Chan, R., Kooijman, E.E., Uppamoochikkal, P., Qiang, W., Weliky, D.P. and Nagle, J.F. 2010. HIV Fusion Peptide Penetrates, Disorders, and Softens T-cell Membrane Mimics. J. Mol. Biol. 402:139-153.
Greenwood , A. I., Pan , J., Mills, T. T., Nagle, J. F., Epand, R. M. and Tristram-Nagle, S. 2008. CRAC Motif Peptide of the HIV-1 gp41Protein Thins SOPC Membranes and Interacts with Cholesterol . Biochim. Biophys. Acta 1778:1120-1130.
Tristram-Nagle, S. and Nagle, J.F. 2007. HIV-1 Fusion Peptide Decreases Bending Energy, Promotes Curved Fusion Intermediates. Biophysical Journal 93:2048-2055.
Submitted by: Stephanie Tristram-Nagle, Carnegie Mellon University
and Kathy Dedrick, CHESS, Cornell University
06/12/2015
