X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
The Macromolecular Diffraction Facility at the Cornell High Energy Synchrotron Source (MacCHESS) held its sixth highly successfully BioSAXS Essentials workshop from May 13th to 16th, 2016. Thirty students from fourteen different institutions (including ones as far away as UC Irvine and the University of Puerto Rico) attended the workshop in person, and fifteen students from eleven institutions and companies attended the course remotely via WebEx and YouTube Live. Five different expert instructors gave students a day and a half of lectures and hands-on tutorials in SAXS fundamentals, data collection, and data processing. On-site students then had two and a half days of hands-on data collection at two different MacCHESS beamlines (G1 and F1), allowing them to obtain valuable practical experience and collect data from research samples.

Dr. Richard Gillilan giving the opening lecture of the BioSAXS Essentials 6 workshop. There were thirty students on site for the workshop.
The workshop kicked off on the 13th with an overview of SAXS and fundamental scattering principles by Dr. Richard Gillilan (MacCHESS), followed by an extremely thorough presentation on best practices for preparing samples for SAXS experiments by Dr. Kushol Gupta (UPENN Medical School). Dr. Gillilan closed out the morning with a two-part lecture covering the basics of SAXS data collection and analysis, with a focus on assessing/ensuring data quality at the beamline.
After lunch (and, perhaps more importantly, coffee) on the 13th, Dr. Thomas Grant (Hauptman-Woodward Institute) gave the first of his two talks on advanced processing methods. He first provided a summary of the processing Dr. Gillilan discussed in the morning session, and then focused on inverse Fourier transform methods and bead model reconstructions from SAXS data. This was followed by a discussion on best publication practices by Dr. Gillilan, which provided crucial information on how to write up and publish SAXS data and models.

Dr. Ileana González-González (Universidad del Turabo, Gurabo, Puerto Rico) prepares her sample for a SEC-SAXS experiment. Students, in groups of 2-4, got 6 hours of beamtime at stations G1 or F1 to obtain hands-on experience and training with Dr. Gillilan and Dr. Hopkins. They were able to collect data on standards and research samples.
The final lectures of the day were also on advanced measurement and processing techniques. The first of these was given by Dr. Steve Meisburger (Princeton), and covered the use of Size Exclusion Chromatography coupled SAXS (SEC-SAXS), including advanced techniques for deconvolving poorly-separated SEC-SAXS data developed at CHESS by the Ando group (Princeton). Dr. Gupta gave his second talk of the day, discussing the analysis of mixtures, flexibility, atomistic modeling, and contrast matching/SANS experiments. Finally, Dr. Grant closed out the day with an overview of rigid body modeling, hybrid techniques, and time resolved SAXS at synchrotrons and XFELs.
The morning of the 14th was devoted to hands-on data processing tutorials, run by Dr. Jesse Hopkins (MacCHESS) and Dr. Meisburger. The first tutorial covered the basics of data processing and verification at the beamline using RAW (currently supported/developed by Dr. Hopkins). The second tutorial walked students through using advanced modeling techniques, including finding the P(r) function with GNOM and using DAMMIF to reconstruct bead models from measured scattering profiles.

Dr. Arnab Modak (University of Connecticut) loads his sample by hand into the sample cell at G1.
During the night of the 13th, and around the clock from noon on the 14th to 5 pm on the 16th students collected SAXS data at the G1 and F1 MacCHESS beamlines with guidance and training from Dr. Gillilan and Dr. Hopkins (the F1 beamline was converted to SAXS for the workshop). Students collected data in groups of 2-4, with at least 6 hours of data collection per group. They had the option of carrying out either standard robotically assisted SAXS or SEC-SAXS during their beamtime. While standard proteins were available for training, most students collected data on research samples they had brought from their labs.
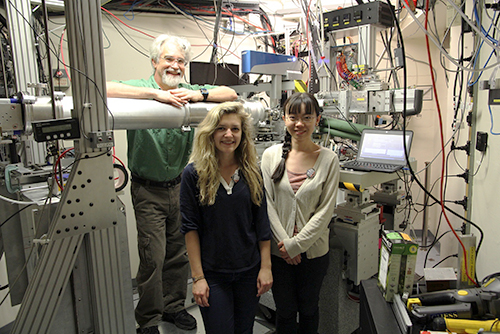
The F1 station is usually a macromolecular crystallography station. In preparation for the course, it was temporarily converted to SAXS use, shown here. Much of the conversion was done by Master’s students Melanie MacMullan (front left) and Manjie Huang (front right) working with Dr. Gillilan (back). Additional work was done by Dr. Hopkins and research support specialists Bill Miller and Scott Smith (not pictured).
Submitted by: Jesse Hopkins, MacCHESS, Cornell University
06/09/2016
