X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
Eddy Arnold of Rutgers University has been studying the HIV virus for a long time. A frequent user of CHESS, Arnold has been using the technique of X-ray crystallography to investigate the structure of HIV proteins and learn more about inhibitors of those proteins which might lead to drugs to fight AIDS. His efforts have been reported here several times (1, 2).
Jeffrey DeStefano, an HIV researcher at the University of Maryland, College Park and post-doc Gauri Nair published a paper in 2008 about their development of a new inhibitor of HIV RT (reverse transcriptase), the viral enzyme which copies viral RNA into DNA in order that it might be incorporated into the host cell's chromosomes by another viral enzyme, integrase (3). The RT inhibitor consisted of a 38 base pair piece of DNA with a particular sequence which binds tightly to the enzyme. Such a specific piece of DNA is called an aptamer.
Arnold and DeStefano then collaborated to study the aptamer using structural biology. Not only did they manage to solve the structure of the RT-aptamer complex, they found that the new inhibitor bound so tightly to RT that it stabilizes the complex and helps it to form better crystals than were previously available (4). Before this, researchers trying to crystallize RT in order to study it with X-ray crystallography had to either chemically cross-link the enzyme to its substrate, or co-crystallize it with an antibody fragment. The new system is not only easier, it provides better results. The X-ray structure was solved at the CHESS F1 beamline.
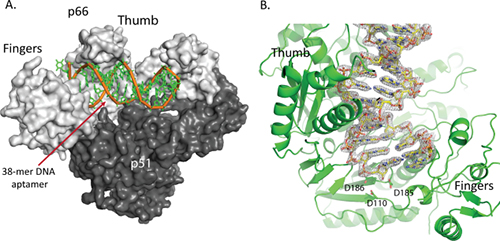
With the RT-aptamer complex in hand, Arnold and DeStefano had a new tool to study the HIV RT enzyme, and they put it to use in solving structures of the enzyme-aptamer complex in various states (5). By adding calcium, a metal which discourages the reaction catalyzed by RT, they solved a structure with an unreacted inhibitor AZTTP bound. By adding magnesium instead, they solved a structure of RT with the AZTMP inhibitor (the reaction product of AZTTP) enzymatically linked to the aptamer; but the aptamer binds so tightly to the enzyme that the aptamer does not translocate to the next position in the way it usually does. Another structure includes the inhibitor alpha-CNP, and yet another the inhibitor foscarnet (phosphonoformic acid). The structures produced by this set of experiments provides a treasure trove of information about the enzyme and the way it works, information that can be put to use in designing better inhibitors and hopefully better drugs to fight the HIV virus. Some of the data for these structures was collected at CHESS beamline F1, and some at NSLS beamline X25.
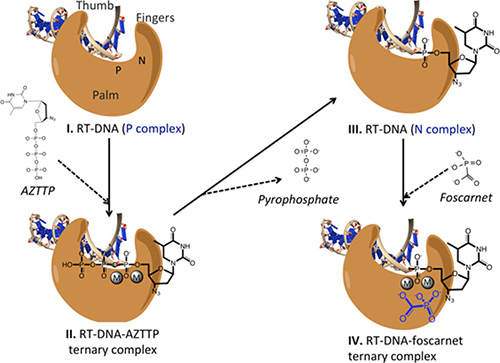
The foscarnet structure is especially intriguing. HIV evolves rapidly, and tends to becomes resistant to drugs over time. Foscarnet is used as an anti-viral therapy for cytomegalovirus and herpes simplex virus infections in some immunocompromised patients. Foscarnet is a weak inhibitor of HIV RT, but it has the curious property that when HIV develops resistance to foscarnet, it regains susceptibility to AZT, a commonly-used drug. Likewise, when HIV develops resistance to AZT, it regains susceptibility to foscarnet. So if foscarnet could be used as the starting point for designing a better inhibitor, that inhibitor would likely make a good complement to the AZT class of drugs. The hope is that the crystallographic structures provided by this research might eventually lead to such advances in AIDS medication.
References:
[1] Mechanism of HIV Resistance to the Drug AZT: mutated reverse transcriptase escapes inactivation by removing AZT that binds to it (2011)
[2] New Insights into anti-AIDS Drug Action (2012)
[3] Novel Aptamer Inhibitors of Human Immunodeficiency Virus Reverse Transcriptase. DeStefano, J.J. and Nair, G.R. Oligonucleotides (2008) 18(2) 133-144.
doi:10.1089/oli.2008.0103
[4] Structure of HIV-1 reverse transcriptase bound to a novel 38-mer hairpin template-primer DNA aptamer. Miller, M. T., Tuske, S., Das, K., DeStefano, J. J. and Arnold, E. (2016) Protein Science, 25: 46-55. doi:10.1002/pro.2776
Structure:
5D3G Structure of HIV-1 Reverse Transcriptase Bound to a Novel 38-mer Hairpin Template-Primer DNA Aptamer
[5] Conformational States of HIV-1 Reverse Transcriptase for Nucleotide Incorporation vs Pyrophosphorolysis-Binding of Foscarnet, Das, K., Balzarini, J., Miller, M.T., Maguire, A.R., DeStefano, J.J., and Arnold, E. (2016), ACS Chem. Biol., 2016, 11 (8), pp 2158-2164
doi: 10.1021/acschembio.6b00187
Structures:
5I42 Structure of HIV-1 Reverse Transcriptase in complex with a DNA aptamer, AZTTP, and CA(2+) ion
5I3U Structure of HIV-1 reverse transcriptase N-site complex; catalytic incorporation OF AZTMP to a DNA aptamer in crystal
5HP1 Structure of HIV-1 reverse transcriptase in complex with a DNA aptamer and foscarnet, a pyrophosphate analog
5HRO Structure of HIV-1 reverse transcriptase in complex with a DNA aptamer and an Alpha-carboxy nucleoside phosphonate inhibitor (alpha-CNP)
Submitted by: David J. Schuller, MacCHESS, Cornell University
09/06/2016
