X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
Since its introduction by Søren Skou (Nielsen) in 2010, the BioXTAS RAW software has been a familiar interface to the many biomedical scientists collecting data at CHESS beamlines in recent years. From the start, RAW was designed specifically with novice users in mind: when scientists arrive at the beamline, they need something fast and easy to learn in the very limited time available … often late at night. The program was literally designed by looking over the shoulders of beamline users as they collected data. But rather than simply create an automated data processing pipeline, we opted to give people the power to fully process data on their own computers at home, if they choose. This allows them to use the same software at other beamlines and even on their own home X-ray sources: from initial raw data reduction to final publication. Indeed, with over 4000 downloads in 2017, RAW is now the primary processing software at several other beamlines and lab source facilities worldwide.
 Richard Gillilan, Jesse Hopkins, and Søren Skou at the annual American Crystallographic Association meeting where they conducted a tutorial in the BioXTAS RAW software. Their new paper describing the software will appear in the latest issue of Journal of Applied Crystallography (open access): scripts.iucr.org/cgi-bin/paper?ge5036
Richard Gillilan, Jesse Hopkins, and Søren Skou at the annual American Crystallographic Association meeting where they conducted a tutorial in the BioXTAS RAW software. Their new paper describing the software will appear in the latest issue of Journal of Applied Crystallography (open access): scripts.iucr.org/cgi-bin/paper?ge5036
RAW is free, open source, and runs on Mac, Windows, and Linux. Jesse Hopkins (MacCHESS) has greatly expanded the capabilities and documentation of the program, making it easier than ever to install at home. RAW can read over 27 different detector image formats and has sophisticated masking and integration capabilities as well as a newly-added algorithm for statistically comparing and merging multiple exposures. Data can be placed on an absolute scale based on water or glassy carbon standards. The program can perform all the customary SAXS analyses such as Guinier analysis, Porod and normalized Kratky plots, and pair distance distribution functions. The program also can serve as an interface for several of the most popular algorithms in the widely used ATSAS suite for solution scattering developed at EMBL.
The most recent and exciting addition to RAW is its ability to untangle mixtures of biomolecules. RAW is specifically designed to work with size exclusion chromatography coupled SAXS (SEC-SAXS), a wildly popular new technique for separating and structurally analyzing mixtures of biomolecules. The software contains the first public implementation of the powerful new Evolving Factor Analysis (EFA) for extracting scattering curves from overlapping peaks in SEC-SAXS chromatograms. The method has already been used successfully by several early external users and promises to be a breakthrough in solving tough separation problems.
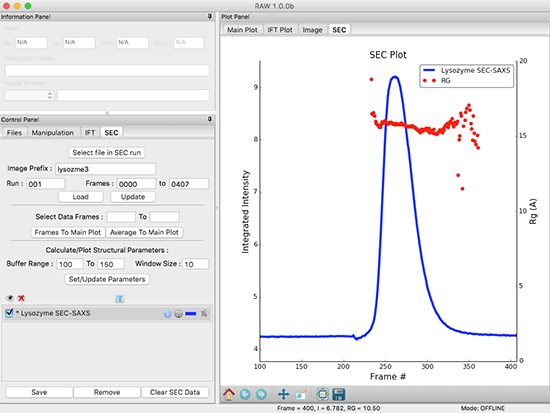 BioXTAS RAW software showing analysis of size exclusion chromatography coupled to small angle X-ray measurements (SEC-SAXS).
BioXTAS RAW software showing analysis of size exclusion chromatography coupled to small angle X-ray measurements (SEC-SAXS).
Full details about RAW including validation tests can be found in the latest issue of the Journal of Applied Crystallography: Hopkins. J. B., Gillilan, R. E., Skou, S. “BioXTAS RAW: improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis.” J. Appl. Cryst. (2017), 50. DOI: 10.1107/S1600576717011438
Open access link: scripts.iucr.org/cgi-bin/paper?ge5036
A complete printable manual, installation packages, HOWTO videos, and tutorials with test data can be found on our Source Forge site: sourceforge.net/projects/bioxtasraw.
The manual, tutorial, installation instructions, and links to the videos are also available as a website at: http://bioxtas-raw.readthedocs.io
Thanks to Jesper Nygaard, Kurt Andersen, Alvin Acerbo, and the many BioSAXS users whose feedback has contributed greatly to RAW. Thanks also to SAXSLAB (saxslab.com)
Submitted by: Richard Gillilan and Jesse Hopkins, MacCHESS, Cornell University
09/14/2017
