X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
Information encoded in our genes controls how we live and grow. As part of this complex process, DNA is transcribed to RNA, one "letter" (nucleotide) at a time, by an enzyme called RNA polymerase (RNAP). At a later stage, 3-nucleotide RNA sequences are translated into protein according to the genetic code.
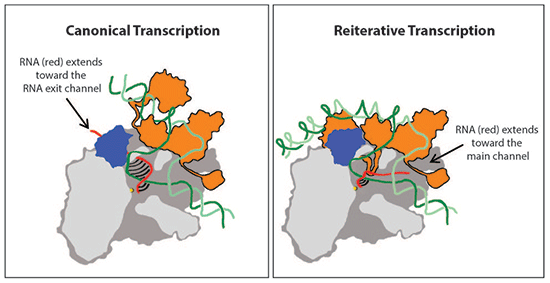
From years of biochemical and structural studies, we have a good understanding of how transcription works - or do we? In "canonical transcription", each nucleotide (A, T, C, or G) in a DNA strand is transcribed into the single complementary nucleotide (U, A, G, or C) in RNA. However, as discovered by Nobel laureate Paul Berg in the 1960's, "reiterative transcription" can also occur, in which a single DNA nucleotide is transcribed into multiple copies of the complementary RNA nucleotide. The latter process, which has important regulatory roles in many species, is not so well understood, and recent work by Katsu Murakami and colleagues at Penn State provides some fascinating insight into how RNAP carries out this type of transcription.
Investigation of the mechanism of action of bacterial RNAP is a major project of the Murakami lab, and crystal structures determined using data from CHESS have been critical to the success of this work. Unlike many macromolecules, some RNAPs can function within a crystal, so that functional intermediate states can be observed. Previous results from the Murakami group have revealed many details of the canonical transcription process. Now, as reported in Proceedings of the National Academy of Sciences (Murakami et al. 2017, PNAS 114, 8211-8216), the group has determined a crystal structure for an RNAP complex caught in the act of reiterative transcription. Data were collected at the F1 station.
The structure (below) shows that, during reiterative transcription, the growing strand of RNA does not extend toward the RNA exit channel (as it does in the canonical case). Instead, it detours towards the "main channel" of RNAP, an action that allows RNA extension without shifting the DNA, so that multiple copies of a single nucleotide are made. The structure also allows determination of the unusual pathway that must be followed when reiterative transcription terminates and normal transcription of the rest of the DNA proceeds.
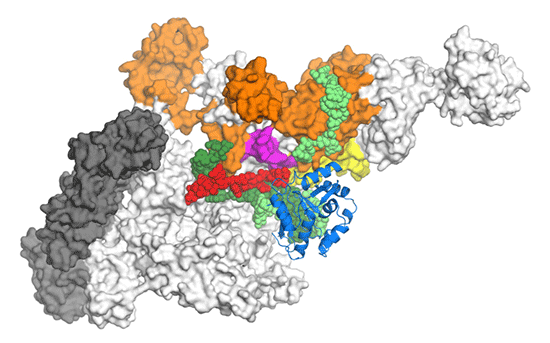 In the figure, RNA is shown in red, DNA in shades of green, and RNAP in gray
(α subunit), white (β' subunit), orange (σ subunit), and blue
(β subunit, part of the domain omitted for clarity).
In the figure, RNA is shown in red, DNA in shades of green, and RNAP in gray
(α subunit), white (β' subunit), orange (σ subunit), and blue
(β subunit, part of the domain omitted for clarity).
The work was highlighted by Penn State in a news release. More information is available at the Murakami group's web site.
Submitted by: Marian Szebenyi, MacCHESS, Cornell University
10/09/2017
