X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
Contact: D. M. Szebenyi (dms35@cornell.edu)
Plant pathogens use "effector proteins" to enhance the infection process in host plants. Plants, in turn, have evolved a defense mechanism in which a pathogen's effector is recognized by a corresponding plant disease resistance protein, and a "hypersensitive response" is triggered, leading to rapid localized cell death which limits the spread of the pathogen. What is the mechanism of recognition? A recent report ("The structural basis for activation of plant immunity by bacterial effector protein AvrPto", W. Xing, Y. Zou, Q. Liu, J. Liu,X. Luo,Q. Huang, S. Chen, L. Zhu, R. Bi, Q. Hao, J.-W. Wu, J.-M. Zhou, and J. Chai, Nature (2007), doi:10.1038/nature06109) describes the crystal structure of a complex of the AvrPto effector from Pseudomonas syringae and the Pto resistance protein from tomato. The AvrPto-Pto interaction is known to trigger a hypersensitive reaction involving cell death initiated by the Prf protein. It was widely thought that binding of AvrPto activates Pto, which then activates Prf by phosphorylating it, but some biochemical evidence suggested that Pto actually inhibits Prf, and that AvrPto relieves this inhibition; structural information about the AvrPto-Pto complex could resolve this issue. A solution structure had been determined for AvrPto alone, but not for the complex.
The crystal structure was determined by a collaboration between workers from the National Institute of Biological Sciences Beijing and MacCHESS; MAD data from a selenomethionine derivative were collected at the Beijing SRF, and structure solution was carried out at at MacCHESS. Initial phases to 4 Å were determined from the MAD data, and refinement was carried out using data to 3.2 Å. The final R-value was 27.1%, with an Rfree of 30.3%. The final model contained the whole AvrPto-Pto complex, except for the first 29 residues of Pto, and included one phosphorylated threonine residue, T199, in Pto.
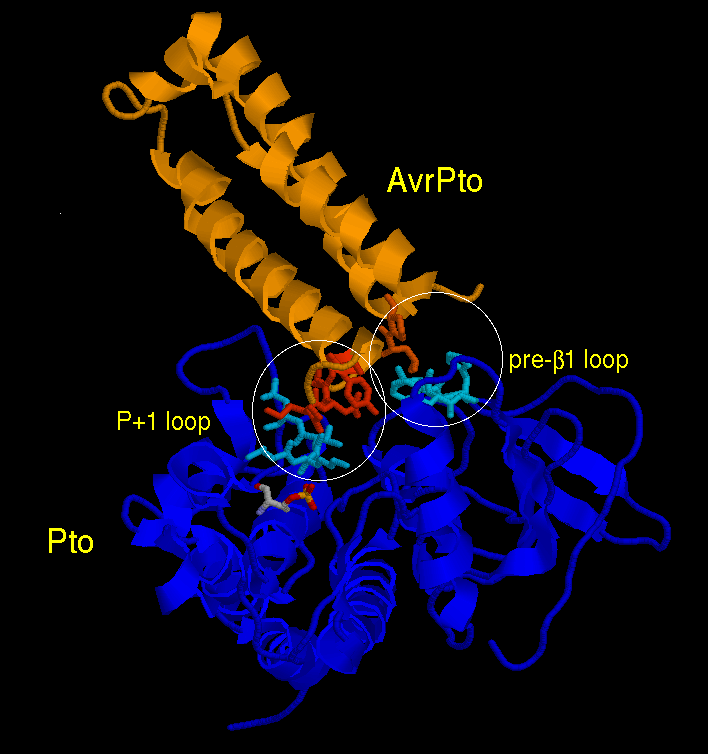
The interaction region between the two proteins involves one end of elongated, mostly-helical, AvrPto and two separate loops on the surface of Pto, designated the "P+1 loop" and the "loop preceding β1". The second interface involves the "GINP" loop (red) of AvrPto, which is disordered in a solution of AvrPto alone but well-ordered in the crystal of the complex. Mutational experiments indicate that both interactions are required for formation of the complex. Comparison of the P+1 loop with the corresponding loops in related proteins, and the phosphorylated condition of T199 (CPK colors), indicate that Pto is in its active state in AvrPto-Pto, and further mutational studies confirm that AvrPto does not bind to inactive Pto.
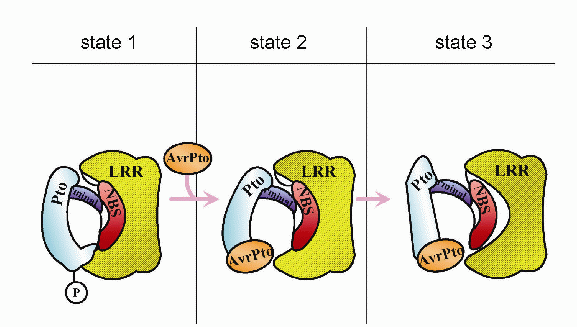
These results suggest that AvrPto binding acts to inhibit the active form of Pto, rather than activating an inactive form. Xing et al. propose the mechanism shown below for the activation of Prf (and hence the hypersensitive reaction): normally (state 1) Prf, comprising N-terminal, LRR and NBS domains, is inactive, and is maintained in that state by interaction with phosphorylated Pto. Binding of AvrPto causes a conformational change in Pto (state 2) which results in "unlocking" and activation of Prf (state 3).
See full article published in Nature
