X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
Polymerase II transcribes DNA to produce messenger RNA. Before the mRNA can be used to produce proteins, it must be modified by the addition of a 7-methyl guanosine "cap", which serves as a recognition site for proteins involved in splicing and translation. The capped protein then binds to CBC, the cap-binding complex, moves out of the nucleus into the cytoplasm, and is released from CBC. Further processing by cytosolic enzymes results in proper splicing and translation into protein. In the nucleus, binding of importin-α to the CBC-RNA complex stabilizes it. In the cytosol, an additional molecule, importin-β binds, destabilizing the CBC-RNA contact and leading to release of mRNA. Importin-β is also involved in nuclear import of CBC. Recent work by the Cerione group (Cornell) has shown how the importins bind to CBC, and why mRNA is released when importin-β binds. The work is reported in: "The Molecular Basis for the Regulation of the Cap-binding Complex by the Importins", S.M.G. Dias, K.F.Wilson, K.S.Rojas, A.L.B. Ambrosio, and R.A. Cerione; Nature Struct. Mol. Biol. Vol. 16, 930-937 (2009)
A 2.2 Å crystal structure of the complex of CBC with an importin-α construct, from which the IBB domain (the flexible N-terminal portion) was deleted, was determined using data collected at CHESS. CBC has two parts: the CBP20 cap-binding domain and the CBP80 regulatory domain. The structure showed that importin-α binds to the N-terminal end of CBP80, which contains two nuclear localization sequences. The crystallization solution also contained a cap analog, which was expected to bind to CBP20. However, the predicted binding site was occupied by an importin-α molecule from a neighboring complex and no cap analog was seen. This observation suggested that release of capped mRNA might be promoted by binding of importin-β at the cap-binding site. Biochemical binding studies with importin-β mutants indicated that residues 45-305 of the protein are involved in binding to CBP20. Models were constructed for the ternary complex using all available structural and biochemical information, and predicted SAXS curves were tested against the measured SAXS data. Good agreement was obtained for the model shown below, in which importin-β blocks access of capped mRNA to the binding site on CBP20. The model is also consistent with biochemical results suggesting that the IBB domain of importin-α (not present in the CBC-importin-α crystal structure) interacts with importin-β.
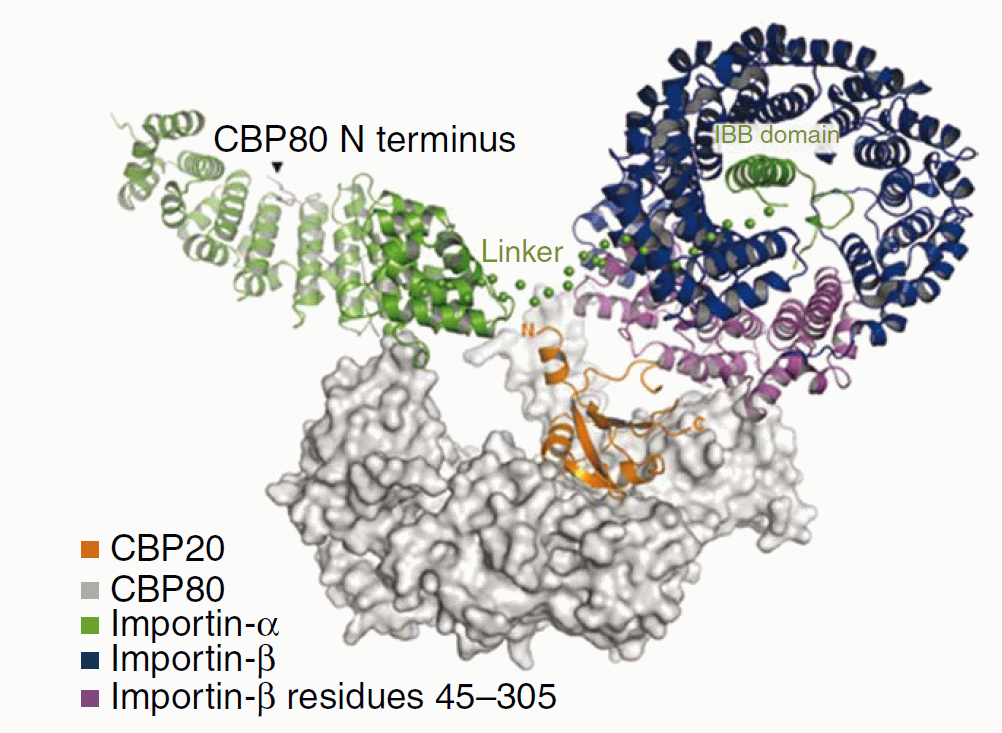
Structural model for CBC-importin-α-importin-β complex. The structure of the CBC (CBP20 and CBP80)-importin-α complex was determined crystallographically (except for the IBB domain, which was deleted to promote crystallization), and importin-β was added using SAXS data, biochemical results, and the known structure of an importin-α-importin-β complex. The location of the IBB domain is unknown; it is probably disordered.
10/27/2009
submitted by: M. Szebenyi
