X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
Many biological processes require the formation of protein complexes containing multiple copies of one or more types of polypeptide chain. It is critical that the protein chains involved correctly recognize each other and assemble into the proper sequence. One mechanism providing high specificity in protein-protein recognition and assembly is domain-swapping, in which structural elements such as β-sheets, which usually occur within a single protein domain, contain chain segments from two different protein molecules. This is generally a symmetric process, so that each member of a group of proteins has part of another's chain inserted into it. There are a number of examples of domain-swapping in natural proteins, but the Loh group (SUNY Upstate Medical University) has now taken a step towards harnessing the mechanism for engineering new structures.
As a test case, the group worked with barnase (Bn), a small, well-ordered, monomeric protein. The plan was to insert another small protein, ubiquitin (Ub), into the sequence of Bn in such a way as to destabilize the native structure. Combination of two or more molecules could then reconstruct an approximation to the original structure.
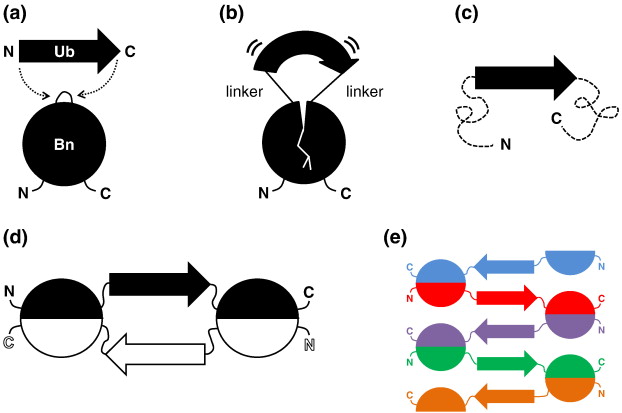
Approach for engineering a domain-swapped complex. a) A ubiquitin (Ub) molecule is inserted into a loop of barnase (Bn). b) Both molecules are stressed, leading (c) to unfolding of Bn. d) A domain-swapped dimer is created which allows both molecules to be folded in near-native conformations. e) A polymeric structure is also possible.
Six different proteins were constructed by protein engineering, with Ub inserted at six different positions along the Bn chain. In order to minimize distortion of the core elements of the structure, the insertions were made in external loop regions. Measurement of molecular size and denaturation parameters indicated that all the constructs formed substantial amounts of dimers or higher oligomers, and all but one were stably folded. To verify that actual domain-swapping was occurring, a 2.2 Å crystal structure was determined, using CHESS data, for the species “BU103” (Ub inserted at position 103 of Bn). As expected, the structure of Bn in the crystal was similar to that of the normal monomer, but with one strand of each molecule's β-sheet coming from another molecule. Probably because of the high concentration of BU103 in the crystallization solution, a polymeric structure, rather than the mixture of dimers and tetramers seen in solution, was found (shown schematically in part (e) of the figure above and in more detail below).
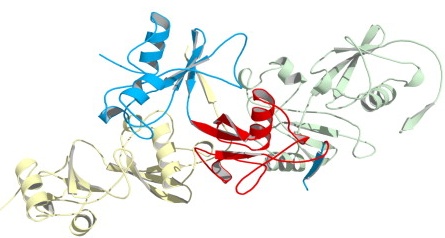
One β-strand of each Bn molecule packs with the β-sheet of another: yellow strand with blue molecule, blue strand with green molecule, etc. The red molecule is Ub.
The Ub conformation was also very similar to its usual one, except at the ends of the chain. These results suggest that it may be possible to use domain-swapping, induced by disruption of a structural element, as a mechanism for self-assembly of oligomers and polymers that retain the function of the component molecules. The research is reported in the Journal of Molecular Biology[1].
Reference:
[1] J.-H.
Ha, J.M. Karchin, N. Walker-Kopp, L.-S. Huang, E.A. Berry, S.N. Loh;
"Engineering Domain-swapped Binding
Interfaces by Mutually Exclusive Folding", J. Mol. Biol.
416, 495-502 (2012)
Submitted by: Marian Szebenyi, MacCHESS, Cornell University
05/10/2012
