X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
RNA polymerase (RNAP) is the molecular Xerox machine that mediates information transfer from DNA to ribosomes for protein synthesis. Usually DNA and RNA polymerases are complex structures that require the presence of multiple protein co-factors. However, in this study Basu and Murakami dissect the reaction mechanism of a single subunit viral RNAP via a time-resolved soak-trigger-freeze X-ray crystallography method [1]. This study provides the final clues to the two-metal-ion reaction mechanism that was first proposed two decades ago by Steitz and Steitz [2].
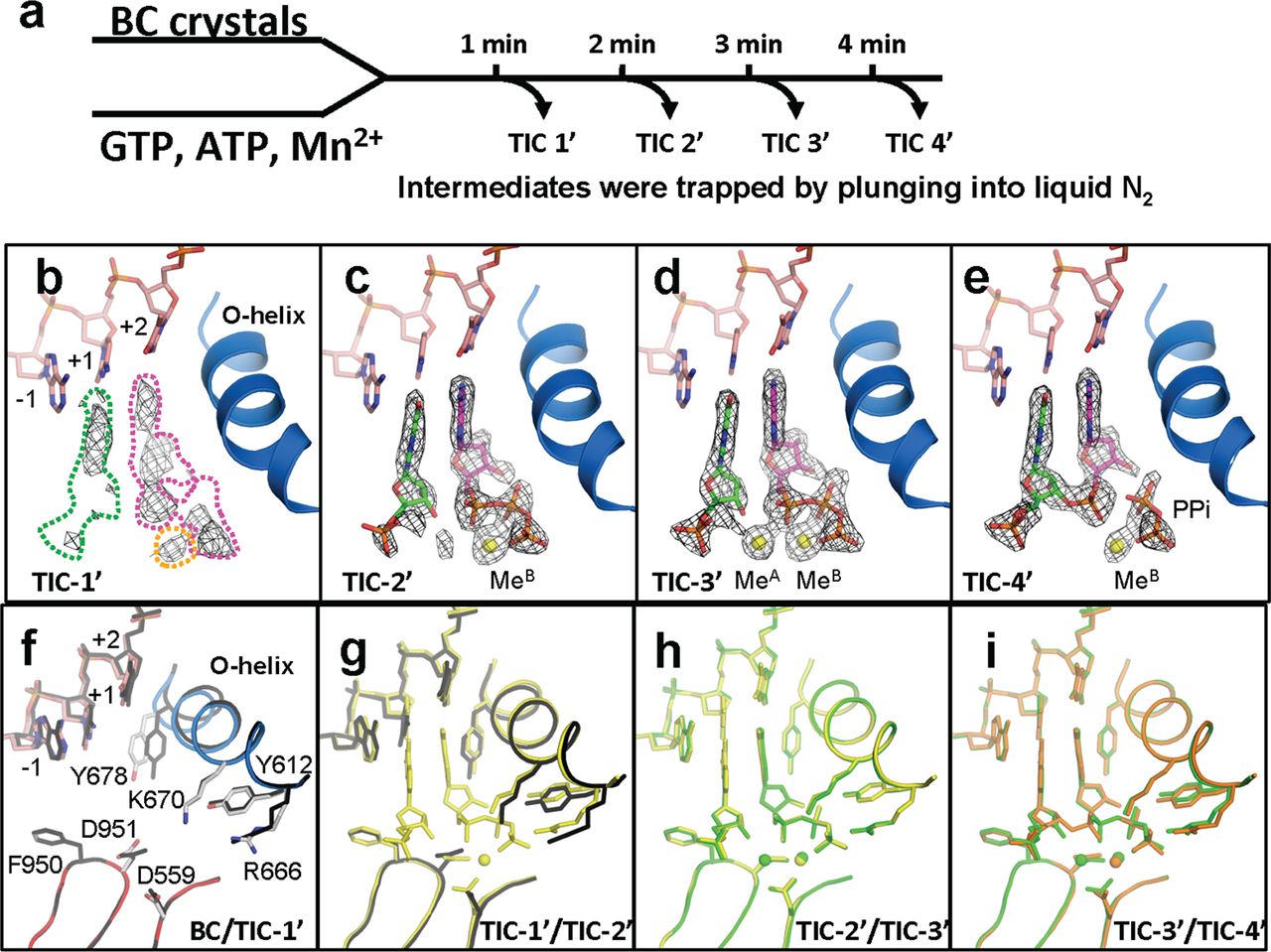
FIGURE 1. Structures of RNAP active site, DNA, nucleotides, and metals during the phosphodiester bond formation. a, scheme of translation initiation complex (TIC) preparations and structure determinations by time-dependent soak-trigger-freeze X-ray crystallography. b–e, electron density showing nucleotides, 2-mer RNA, pyrophosphate, and metals found in the TIC-1′–TIC-4′ structures. f–i, conformational changes of RNAP and DNA during transcription initiation showing superposition of the BC versus TIC-1′ (f), TIC-1′ versus TIC-2′ (g), TIC-2′ versus TIC-3′ (h), and TIC-3′ versus TIC-4′ (i). TIC-1′, TIC-2′, TIC-3′, and TIC-4′ are colored black, yellow, green, and orange, respectively.
In this study Pennsylvania State University Biochemistry and Molecular Biology Professor Katsu Murakami and graduate student Ritwika Basu took advantage of enzyme catalyzed reactions in crystallo. Due to the relatively slower reaction rates in tightly packed crystals, they were able to trap the otherwise short-lived reaction intermediates (Figure 1). They first soaked the crystals of RNAP-promoter DNA complex (BC – binary complex) with the respective metal ions and nucleotides, followed by flash-cooling the crystals at different time points. By doing this they were able to track the appearance and disappearance of electron density corresponding to the incoming/outgoing metal ions, and assign the conformational changes of the catalytically essential O-helix to specific events in the reaction scheme (Figure 2, a).
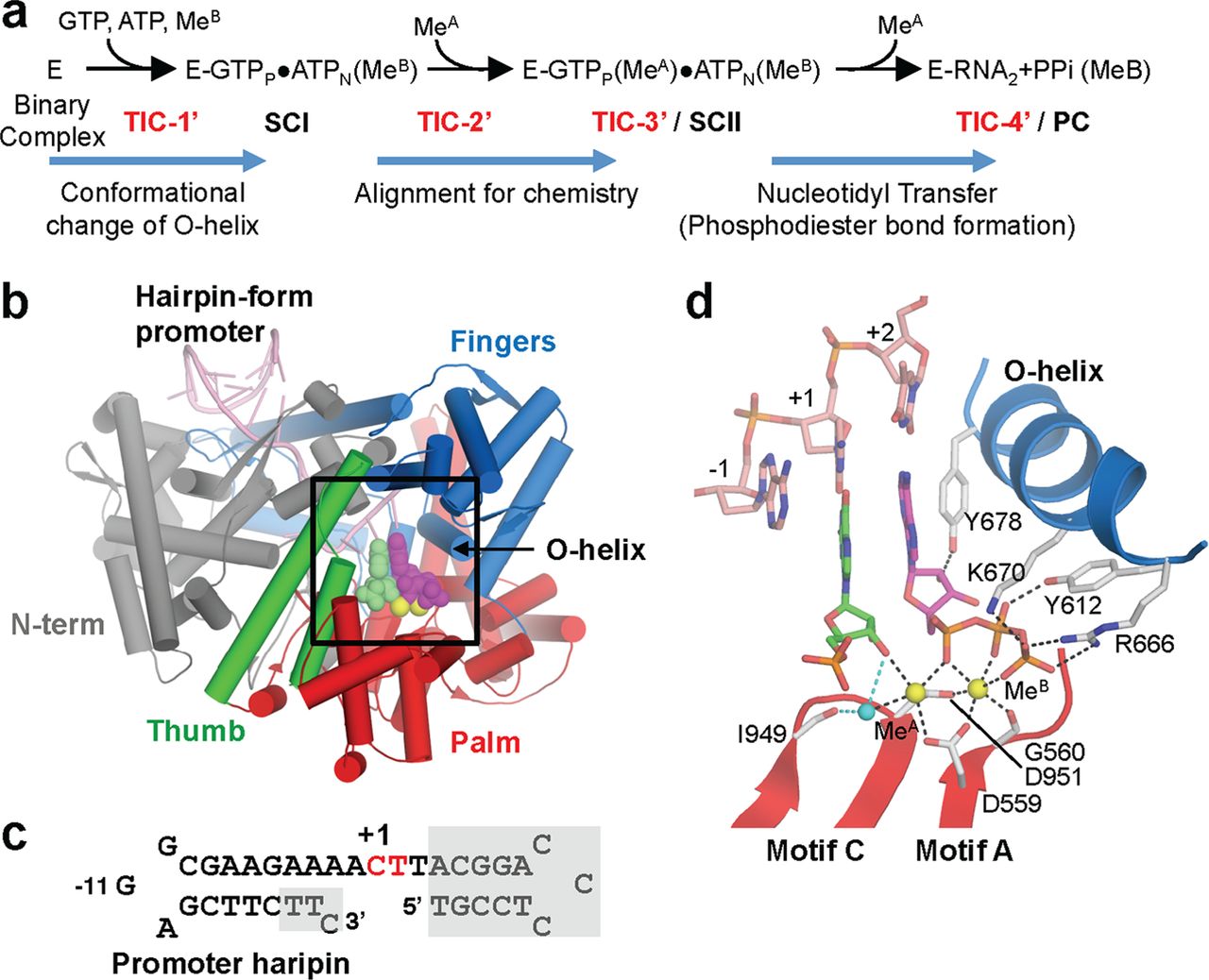
FIGURE 2. Structure of the transcript initiation complex. a, schematic of transcription initiation. The RNAP and promoter DNA complex is depicted as E. The appearance of intermediates, from TIC-1′ to TIC-4′, isolated in this study, as well as SCI, SCII, and PC identified in the previous study [3], are indicated. b, overall structure of TIC-3′. Each domain and the O-helix of RNAP are denoted by a unique color and labeled. c, sequence of the DNA used for crystallization (red, +1 and +2 nucleotide binding sites; gray boxes, disordered in the crystal structures). The position of promoter hairpin is indicated. d, TIC-3′ active site. The amino acid residues involved in nucleotides and metal binding are shown. Nucleotides at positions +1 (green) and +2 (magenta) are shown. Mn2+ and water are depicted by yellow and cyan spheres, respectively.
This study is in good agreement with previous Raman crystallography experiments and provides further evidence of the temporal order of substrate assembly and metal co-factor binding in the active site of the single subunit RNA polymerases [4].
The data for this study was collected at the F1 beamline at CHESS. The findings of this research study were published in The Journal of Biological Chemistry.
References:
- Basu R. S., Murakami K. S. (2013) Watching the bacteriophage N4 RNA polymerase transcription by time-dependent soak-trigger-freeze X-ray crystallography. J Biol Chem. 288(5), 3305-11.
- Steitz T. A., Steitz J. A. (1993) A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. U.S.A. 90, 6498–6502
- Gleghorn M. L., Davydova E. K., Basu R., Rothman-Denes L. B., Murakami K. S. (2011) X-ray crystal structures elucidate the nucleotidyl transfer reaction of transcript initiation using two nucleotides. Proc. Natl. Acad. Sci. U.S.A. 108, 3566–3571
- Chen Y., Basu R., Gleghorn M. L., Murakami K. S., Carey P. R. (2011) Time-resolved events on the reaction pathway of transcript initiation by a single-subunit RNA polymerase: Raman crystallographic evidence. J. Am. Chem. Soc. 133, 12544–12555
Submitted by: Tiit Lukk, MacCHESS, Cornell University
7/15/2013
