X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
The acronym LOV stands for light, oxygen, or voltage – these are stimuli that can act on a LOV protein domain to produce a physiological response; this is one form of signal transduction. In the phototropic response of plants, for example, the amount of blue light shining on a leaf regulates the size of its stomatal apertures (stomata are vent holes for gas/water vapor). Another example is found in bacteria, where oxygen sensors containing LOV domains have been shown to regulate nitrogen fixation. LOV domains are structural homologues of the PAS superfamily, a group of diverse sensory proteins. Each LOV domain contains a non-covalently bound flavin mononucleotide (FMN) that performs reversible photochemistry [1]. The first X-ray crystal structure of a plant LOV domain was solved more than a decade ago by the Moffat group. That study answered many important questions regarding the binding geometry of the FMN cofactor and the possible course of the photochemical reaction [2]. In a more recent study, Cornell University professor Brian Crane and colleagues dissected the mechanism of light-induced subunit dissociation of RsLOV [3], a LOV domain photoreceptor from the photosynthetic Gram-negative bacteria Rhodobacter sphaeroides (Figure 1).
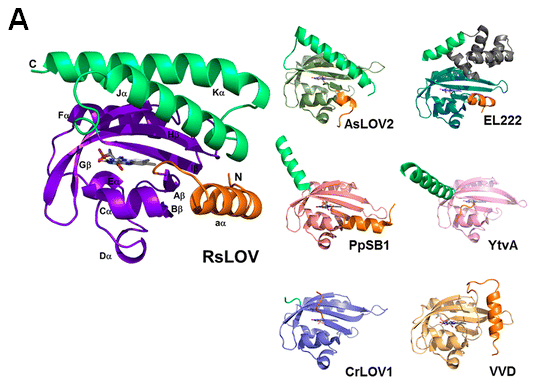
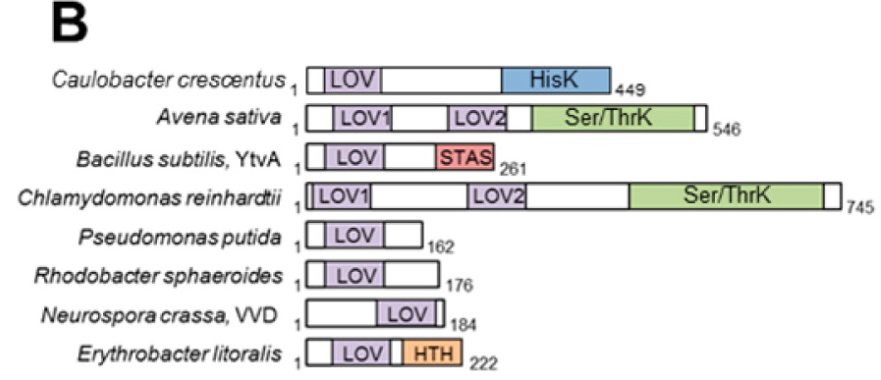
FIGURE 1. Diversity of enzymes containing the LOV domain. A) LOV subunit structures. RsLOV structure in the dark state; dark-state structures of A. sativa LOV2, E. litoralis EL222, P. putida PpSB1 LOV, B. subtilis YtvA LOV, C. reinhardtii LOV1, and N. crassa VVD; N-terminal extensions are colored orange and C-terminal extensions light green. B) Domain arrangement of sequence-aligned LOV domain proteins. Abbreviations: HisK, histidine kinase; Ser/ThrK, serine/threonine kinase; STAS, sulfate transporter anti-s antagonist; HTH, helix−turn−helix.
In this study, the Crane group explored the “photodissociation” mechanism of RsLOV via the avenues of site-directed mutagenesis, size-exclusion chromatography, X-ray crystallography, SAXS, multi-angle light scattering, and time-resolved absorption spectroscopy. This multifaceted approach enabled them to identify key residues involved in the process, as well as to obtain the decay kinetics for the covalent cysteine-FMN adduct that triggers the conformational change. Utilizing small-angle X-ray scattering, they were able to not only validate the transition between the dark-state dimer and the light-induced monomeric form of RsLOV, but also provide visual proof of that switch in the form of a scattering envelope model (Figure 2).
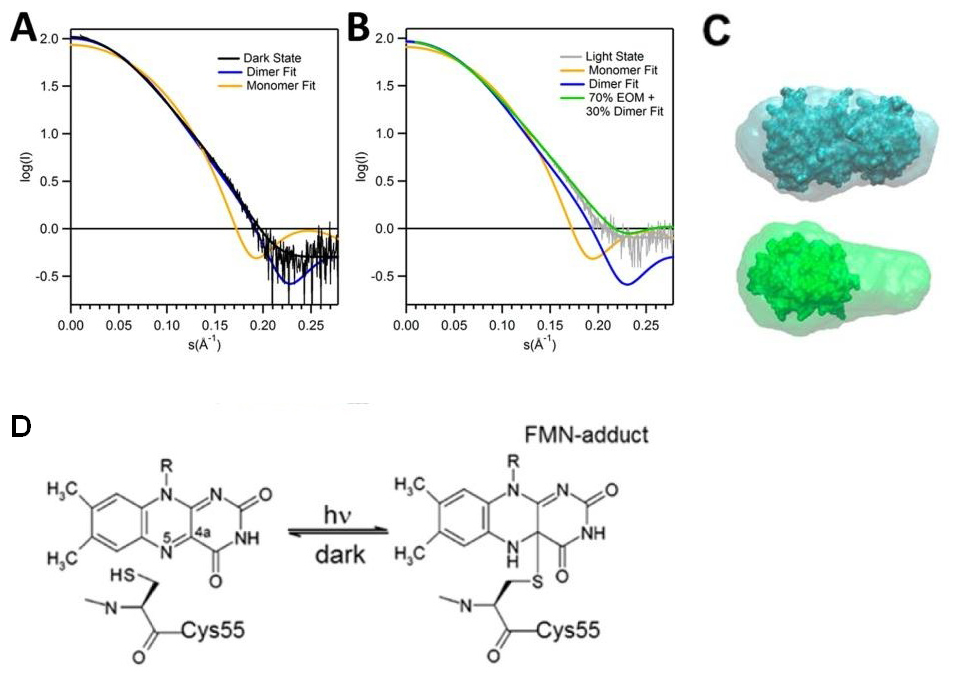
FIGURE 2. SAXS of RsLOV in dark and light states. Fits of theoretical monomer and dimer scattering curves for dark-state and (A) light-state (B). The best fit to the light-state experimental data was obtained using the ensemble optimization method (lime). C) Scattering envelope models of dark-state RsLOV (top) and light-state (bottom) RsLOV, superimposed with the crystal structures as a complete dimer or truncated to a monomer. D) Schematic of light-induced cysteinyl-FMN (C4a) covalent adduct, the trigger for LOV domain monomerization.
X-ray diffraction and SAXS data were collected at CHESS on the A1 and F2 beamlines, respectively.
References:
- Christie J. M., Salomon M., Nozue K., Wada M., Briggs W.R. (1999) LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc Natl Acad Sci U S A. 96(15), 8779-83.
- Crosson S, Moffat K. (2001) Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc Natl Acad Sci U S A. 98(6), 2995-3000.
- Conrad K.S., Bilwes A.M., Crane B.R. (2013) Light-induced subunit dissociation by a light-oxygen-voltage domain photoreceptor from Rhodobacter sphaeroides. Biochemistry 52(2), 378-91.
Submitted by: Tiit Lukk, MacCHESS, Cornell University
9/17/2013
