X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
Each cell contains a copy of one’s genome encoded in the form of a double-stranded DNA molecule. If laid out in a linear fashion, it would reach several feet in length. Eukaryotic cells have found a nifty way of compacting this long strand into a more manageable size so that a cell can easily contain it. By wrapping portions of the DNA around histone octamers, the primary level of compactness is reached, which forms the basic building block of chromatin. Access to any portion of the DNA strand is ensured by continual remodeling of the chromatin structure, whereby small access sites are created and closed shortly after – just long enough for the cell to read that portion of the genetic code.
The complex mechanism of chromatin remodeling has received a lot of attention as dysfunction has been linked to a range of diseases, including cancer, cardiomyopathies, and neurological developmental disorders. Turegun, et al [1] sought to shed a light on a particular family of remodelers, the SWI/SNF family of remodelers. Although the recruitment of several actin-related proteins by SWI/SNF, namely Arp7 and Arp9, is well understood, the role of the recently discovered subunit Rtt102 is unknown. The group used a combination of biochemical assays, circular dichroism and small-angle x-ray scattering to further investigate potential interactions between Rtt102 and the conformation of SWI/SNF and recruitment of actin-related proteins.
Bengi Turegun, a PhD candidate in Biochemistry and Molecular Biophysics at The University of Pennsylvania, took part in the BioSAXS Essentials course held in March 2013. During this 3-day course participants attend several BioSAXS lectures on the fundamentals of BioSAXS as well as understanding how to interpret and analyze data. Several advanced topics are also covered for those interested. The course also includes several hours of individual hands-on experience at MacCHESS BioSAXS beamlines, and attendees often bring their own samples to work with.
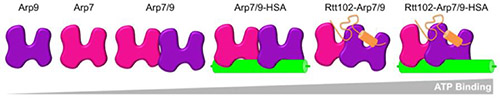
The group used data collected during the course at station F2 to establish monodispersity and calculate pair distribution functions of several complexes involving Rtt102, Arp7, and Arp9. By calculating low-resolution envelopes of two Rtt102 containing complexes and comparing the pair distribution functions of the Arp7/9-ATPase complex, with and without Rtt102, they were able to determine that Rtt102 is not required for the formation of a proper Arp7/9-ATPase complex, but that Rtt102 does result in a more compact and stable conformation. Based on their findings, Rtt102 serves mostly as a stabilizing factor and regulator as it stabilizes and increases the nucleotide-binding affinity of the Rtt102-Arp7/9-HSA complex (HSA is a domain of the SWI/SNF remodeler; see figure).
References:
[1] B. Turegun, D.J. Kast, R. Dominquez: "Subunit Rtt102 Controls the Conformation of the Arp7/9 Heterodimer and Its Interactions with Nucleotide and the Catalytic Subunit of SWI/SNF Remodelers", J Biol Chem 288(50): 35758:35768 (2013).
Submitted by: Alvin Acerbo, MacCHESS, Cornell University
02/07/2014
