X-RAY RUNS: Apply for Beamtime
2017 Nov 1 - Dec 21
2018 Feb 7 - Apr 3
2018 Proposal/BTR deadline: 12/1/17
2018 Apr 11 - Jun 4
2018 Proposal/BTR deadline: 2/1/18
Proteases, enzymes that cleave proteins, are found both free-floating and embedded in membranes. Reactions involving the former are well understood, but the workings of the latter have remained mysterious – how are reactions controlled inside the viscous, two-dimensional membrane, from which water is excluded? Insight into this question has biomedical significance, as membrane proteases are required for the cleavage of transmembrane anchors to release signaling proteins from the membrane, and disruption of this process is implicated in more than a dozen diseases. For example, the intramembrane rhomboid proteases are implicated in Parkinson's disease and parasite invasion.
Structural studies of rhomboid proteases, in particular E. coli GlpG, have revealed the molecular architecture of these enzymes and led to a model for how they work [1]. However, the model was based on extrapolations from soluble proteases, due to the difficulty of monitoring chemical reactions within a membrane. Recently the Urban group (Johns Hopkins) has developed an inducible reconstitution system with which the kinetics of proteolysis by GlpG can be studied in real time, within a membrane. To avoid premature cleavage of substrate during the reconstitution process, it was necessary to have a way to turn proteolysis off (during reconstitution) and on (during kinetic measurements). A pH change does this, but it was necessary to consider what it might do to the protein structure. Hence, the group used CHESS F1 to determine the structures of a truncated form of GlpG, to 2.4 Å resolution, at pH 4.5 and pH 7.5, using molecular replacement for phasing. The two structures (Figure 1) were nearly identical, with one critical difference in an active site serine residue, which explains why the enzyme is active at pH 7.5 but not at pH 4.5.
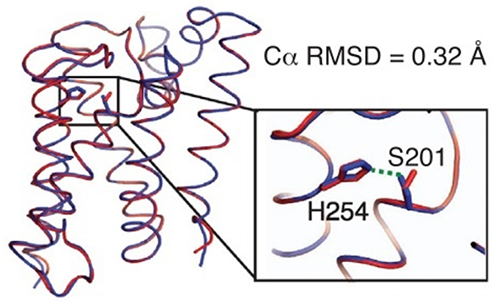
Figure 1. A small difference between the low pH (red) and neutral pH (blue) structures controls the activity of the GlpG protease.
Assured that pH change would have no extraneous effects on enzyme structure, the group proceeded to measure rhomboid protease kinetics in the membrane environment. Unexpectedly, the large differences in proteolytic efficiency among various proteases and substrates turned out to be determined by accessibility of the active site by the substrate, not by enzyme-substrate affinity. Intramembrane rhomboid proteolysis is a slow reaction controlled by the kinetics of substrate approach through the membrane. This behavior is unlike other studied proteases but similar to that of some DNA-repair enzymes; it has important implications for drug design.
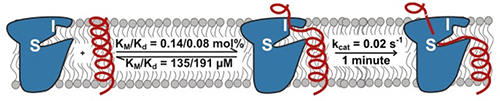
Figure 2. Model for rhomboid protease function in a membrane. I, interrogation site; S, scission site.
References:
[1] S. Urban, “Taking the plunge: integrating structural, enzymatic and computational insights into a unified model for membrane-immersed rhomboid proteolysis”, Biochem. J. 425, 501-512 (2010).
[2] S.W. Dickey, R.P. Baker, S. Cho, S. Urban, “Proteolysis inside the membrane is a rate-governed reaction not driven by substrate affinity”, Cell 155, 1270-1281 (2013).
Submitted by: Marian Szebenyi, MacCHESS, Cornell University
08/13/2014
